Analysis of pmpD expression and PmpD post-translational processing during the life cycle of Chlamydia trachomatis serovars A, D, and L2
- PMID: 19367336
- PMCID: PMC2666266
- DOI: 10.1371/journal.pone.0005191
Analysis of pmpD expression and PmpD post-translational processing during the life cycle of Chlamydia trachomatis serovars A, D, and L2
Abstract
Background: The polymorphic membrane protein D (PmpD) in Chlamydia is structurally similar to autotransporter proteins described in other bacteria and may be involved in cellular and humoral protective immunity against Chlamydia. The mechanism of PmpD post-translational processing and the role of its protein products in the pathogenesis of chlamydial infection have not been very well elucidated to date.
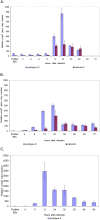
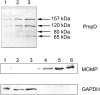
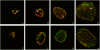
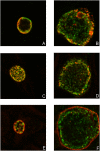
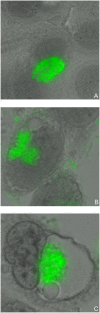
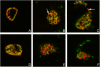
Methodology/principal findings: Here we examined the expression and post-translational processing of the protein product of the pmpD gene during the life cycle of C. trachomatis serovars A, D, and L2. Each of these three serovars targets different human organs and tissues and encodes a different pmpD gene nucleotide sequence. Our quantitative real-time reverse transcription polymerase chain reaction results demonstrate that the pmpD gene is up-regulated at 12-24 hours after infection regardless of the Chlamydia serovar. This up-regulation is coincidental with the period of exponential growth and replication of reticulate bodies (RB) of Chlamydia and indicates a probable similarity in function of pmpD in serovars A, D, and L2 of Chlamydia. Using mass spectrometry analysis, we identified the protein products of post-translational processing of PmpD of C. trachomatis serovar L2 and propose a double pathway model for PmpD processing, with one cleavage site between the passenger and autotransporter domains and the other site in the middle of the passenger domain. Notably, when Chlamydia infected culture cells were subjected to low (28 degrees C) temperature, PmpD post-translational processing and secretion was found to be uninhibited in the resulting persistent infection. In addition, confocal microscopy of cells infected with Chlamydia confirms our earlier hypothesis that PmpD is secreted outside Chlamydia and its secretion increases with growth of the chlamydial inclusion.
Conclusion/significance: The results of this current study involving multiple Chlamydia serovars support the general consensus that the pmpD gene is maximally expressed at mid infection and provide new information about PmpD as an autotransporter protein which is post-translationally processed and secreted outside Chlamydia during normal and low temperature induced persistent chlamydial infection.
Conflict of interest statement
Figures




References
-
- Stephens RS, Kalman S, Lammel C, Fan J, Marathe R, et al. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science. 1998;282(5389):754–9. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

