The wheat omega-gliadin genes: structure and EST analysis
- PMID: 19367421
- PMCID: PMC2700870
- DOI: 10.1007/s10142-009-0122-2
The wheat omega-gliadin genes: structure and EST analysis
Abstract
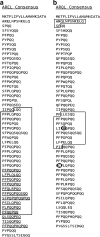
A survey and analysis is made of all available omega-gliadin DNA sequences including omega-gliadin genes within a large genomic clone, previously reported gene sequences, and ESTs identified from the large wheat EST collection. A contiguous portion of the Gli-B3 locus is shown to contain two apparently active omega-gliadin genes, two pseudogenes, and four fragments of the 3' portion of omega-gliadin sequences. Comparison of omega-gliadin sequences allows a phylogenetic picture of their relationships and genomes of origin. Results show three groupings of omega-gliadin active gene sequences assigned to each of the three hexaploid wheat genomes, and a fourth group thus far consisting of pseudogenes assigned to the A-genome. Analysis of omega-gliadin ESTs allows reconstruction of two full-length model sequences encoding the AREL- and ARQL-type proteins from the Gli-A3 and Gli-D3 loci, respectively. There is no DNA evidence of multiple active genes from these two loci. In contrast, ESTs allow identification of at least three to four distinct active genes at the Gli-B3 locus of some cultivars. Additional results include more information on the position of cysteines in some omega-gliadin genes and discussion of problems in studying the omega-gliadin gene family.
Figures
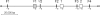





References
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1016/j.jcs.2007.02.001', 'is_inner': False, 'url': 'https://doi.org/10.1016/j.jcs.2007.02.001'}]}
- Altenbach SB, Kothari KM (2007) Omega gliadin genes expressed in Triticum aestivum cv. Butte 86: effects of post-anthesis fertilizer on transcript accumulation during grain development. J Cereal Sci 46:169–177
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1007/BF00020565', 'is_inner': False, 'url': 'https://doi.org/10.1007/bf00020565'}, {'type': 'PubMed', 'value': '1893105', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/1893105/'}]}
- Anderson OD (1991) Characterization of members of a pseudogene subfamily of the wheat alpha-gliadin storage protein genes. Plant Mol Biol 16:335–337 - PubMed
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1007/s001220050531', 'is_inner': False, 'url': 'https://doi.org/10.1007/s001220050531'}]}
- Anderson OD, Litts JC, Greene FC (1997) The α-gliadin gene family. I. Characterization of ten new wheat α-gliadin gene genomic clones, evidence for limited sequence conservation of flanking DNA, and Southern analysis of the gene family. Theor Appl Genet 95:50–58
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'PubMed', 'value': '12590343', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/12590343/'}]}
- Anderson OD, Rausch C, Moullet O, Lagudah E (2003) The wheat D-genome HMW-glutenin locus: BAC sequencing, gene distribution, and retrotransposon clusters. Funct Integr Genomics 3:56–68 - PubMed
-
- Anderson OD, Carollo V, Chao S, Laudencia-Chingcuanco D, Lazo GR (2004) The use of ESTs to analyze the spectrum of wheat seed proteins. In: The Gluten Proteins. Proceedings 8th Gluten Workshop, pp 18–21
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

