Characterization and comprehensive proteome profiling of exosomes secreted by hepatocytes
- PMID: 19367702
- PMCID: PMC2696236
- DOI: 10.1021/pr8004887
Characterization and comprehensive proteome profiling of exosomes secreted by hepatocytes
Abstract
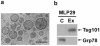
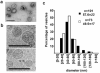
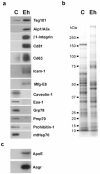
Exosomes represent a discrete population of vesicles that are secreted from various cell types to the extracellular media. Their protein and lipid composition are a consequence of sorting events at the level of the multivesicular body, a central organelle which integrates endocytic and secretory pathways. Characterization of exosomes from different biological samples has shown the presence of common as well as cell-type specific proteins. Remarkably, the protein content of the exosomes is modified upon pathological or stress conditions. Hepatocytes play a central role in the body response to stress metabolizing potentially harmful endogenous substances as well as xenobiotics. In the present study, we described and characterized for the first time exosome secretion in nontumoral hepatocytes, and with the use of a systematic proteomic approach, we establish the first extensive proteome of a hepatocyte-derived exosome population which should be useful in furthering our understanding of the hepatic function and in the identification of components that may serve as biomarkers for hepatic alterations. Our analysis identifies a significant number of proteins previously described among exosomes derived from others cell types as well as proteins involved in metabolizing lipoproteins, endogenous compounds and xenobiotics, not previously described in exosomes. Furthermore, we demonstrated that exosomal membrane proteins can constitute an interesting tool to express nonexosomal proteins into exosomes with therapeutic purposes.
Figures


References
-
- Fevrier B, Raposo G. Exosomes: endosomal-derived vesicles shipping extracellular messages. Current Opinion in Cell Biology. 2004;16(4):415–421. - PubMed
-
- Johnstone RM. Revisiting the road to the discovery of exosomes. Blood Cells Mol Dis. 2005;34(3):214–9. - PubMed
-
- van Niel G, Porto-Carreiro I, Simoes S, Raposo G. Exosomes: a common pathway for a specialized function. J Biochem (Tokyo) 2006;140(1):13–21. - PubMed
-
- Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, Schwille P, Brugger B, Simons M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 2008;319(5867):1244–7. - PubMed
-
- Novikoff AB, Essner E, Quintana N. Golgi Apparatus and Lysosomes. Fed Proc. 1964;23:1010–22. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

