Studying bacteria in respiratory specimens by using conventional and molecular microbiological approaches
- PMID: 19368727
- PMCID: PMC2678980
- DOI: 10.1186/1471-2466-9-14
Studying bacteria in respiratory specimens by using conventional and molecular microbiological approaches
Abstract
Background: Drawing from previous studies, the traditional routine diagnostic microbiology evaluation of samples from chronic respiratory conditions may provide an incomplete picture of the bacteria present in airways disease. Here, the aim was to determine the extent to which routine diagnostic microbiology gave a different assessment of the species present in sputa when analysed by using culture-independent assessment.
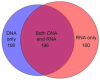
Methods: Six different media used in routine diagnostic microbiology were inoculated with sputum from twelve patients. Bacterial growth on these plates was harvested and both RNA and DNA extracted. DNA and RNA were also extracted directly from the same sample of sputum. All nucleic acids served as templates for PCR and reverse transcriptase-PCR amplification of "broad range" bacterial 16S rRNA gene regions. The regions amplified were separated by Terminal Restriction Fragment Length Polymorphism (T-RFLP) profiling and compared to assess the degree of overlap between approaches.
Results: A mean of 16.3 (SD 10.0) separate T-RF band lengths in the profiles from each sputum sample by Direct Molecular Analysis, with a mean of 8.8 (SD 5.8) resolved by DNA profiling and 13.3 (SD 8.0) resolved by RNA profiling. In comparison, 8.8 (SD 4.4) T-RF bands were resolved in profiles generated by Culture-derived Molecular Analysis. There were a total of 184 instances of T-RF bands detected in the direct sputum profiles but not in the corresponding culture-derived profiles, representing 83 different T-RF band lengths. Amongst these were fifteen instances where the T-RF band represented more than 10% of the total band volume (with a mean value of 23.6%). Eight different T-RF band lengths were resolved as the dominant band in profiles generated directly from sputum. Of these, only three were detected in profiles generated from the corresponding set of cultures.
Conclusion: Due to their focus on isolation of a small group of recognised pathogens, the use of culture-dependent methods to analyse samples from chronic respiratory infections can provide a restricted understanding of the bacterial species present. The use of a culture-independent molecular approach here identifies that there are many bacterial species in samples from CF and COPD patients that may be clinically relevant.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

