Highly sensitive and high-throughput analysis of plant hormones using MS-probe modification and liquid chromatography-tandem mass spectrometry: an application for hormone profiling in Oryza sativa
- PMID: 19369275
- PMCID: PMC2709547
- DOI: 10.1093/pcp/pcp057
Highly sensitive and high-throughput analysis of plant hormones using MS-probe modification and liquid chromatography-tandem mass spectrometry: an application for hormone profiling in Oryza sativa
Abstract
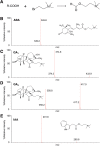
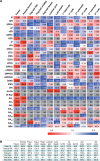
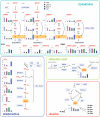
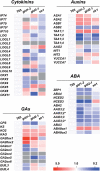
We have developed a highly sensitive and high-throughput method for the simultaneous analysis of 43 molecular species of cytokinins, auxins, ABA and gibberellins. This method consists of an automatic liquid handling system for solid phase extraction and ultra-performance liquid chromatography (UPLC) coupled with a tandem quadrupole mass spectrometer (qMS/MS) equipped with an electrospray interface (ESI; UPLC-ESI-qMS/MS). In order to improve the detection limit of negatively charged compounds, such as gibberellins, we chemically derivatized fractions containing auxin, ABA and gibberellins with bromocholine that has a quaternary ammonium functional group. This modification, that we call 'MS-probe', makes these hormone derivatives have a positive ion charge and permits all compounds to be measured in the positive ion mode with UPLC-ESI-qMS/MS in a single run. Consequently, quantification limits of gibberellins increased up to 50-fold. Our current method needs <100 mg (FW) of plant tissues to determine phytohormone profiles and enables us to analyze >180 plant samples simultaneously. Application of this method to plant hormone profiling enabled us to draw organ distribution maps of hormone species in rice and also to identify interactions among the four major hormones in the rice gibberellin signaling mutants, gid1-3, gid2-1 and slr1. Combining the results of hormone profiling data with transcriptome data in the gibberellin signaling mutants allows us to analyze relationships between changes in gene expression and hormone metabolism.
Figures
References
-
- Bai F, DeMason D. Hormone interactions and regulation of Unifoliata, PsPK2, PsPIN1 and LE gene expression in pea (Pisum sativum) shoot tips. Plant Cell Physiol. 2006;47:935–948. - PubMed
-
- Chiwocha SD, Abrams SR, Ambrose SJ, Cutler AJ, Loewen M, Ross AR, et al. A method for profiling classes of plant hormones and their metabolites using liquid chromatography–electrospray ionization tandem mass spectrometry: an analysis of hormone regulation of thermodormancy of lettuce (Lactuca sativa L.) seeds. Plant J. 2003;35:405–417. - PubMed
-
- Chow B, McCourt P. Plant hormone receptors: perception is everything. Genes Dev. 2006;20:1998–2008. - PubMed
-
- Davies PJ, editor. Plant Hormones. Dordrecht: Kluwer Academic Publishers; 2004. Biosynthesis, Signal Transduc-tion, Action.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

