Herpes simplex virus glycoprotein B associates with target membranes via its fusion loops
- PMID: 19369321
- PMCID: PMC2698560
- DOI: 10.1128/JVI.00301-09
Herpes simplex virus glycoprotein B associates with target membranes via its fusion loops
Abstract
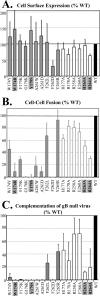
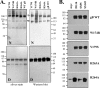
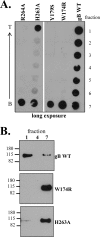
Herpes simplex virus (HSV) glycoproteins gB, gD, and gH/gL are necessary and sufficient for virus entry into cells. Structural features of gB are similar to those of vesicular stomatitis virus G and baculovirus gp64, and together they define the new class III group of fusion proteins. Previously, we used mutagenesis to show that three hydrophobic residues (W174, Y179, and A261) within the putative gB fusion loops are integral to gB function. Here we expanded our analysis, using site-directed mutagenesis of each residue in both gB fusion loops. Mutation of most of the nonpolar or hydrophobic amino acids (W174, F175, G176, Y179, and A261) had severe effects on gB function in cell-cell fusion and null virus complementation assays. Of the six charged amino acids, mutation of H263 or R264 also negatively affected gB function. To further analyze the mutants, we cloned the ectodomains of the W174R, Y179S, H263A, and R264A mutants into a baculovirus expression system and compared them with the wild-type (WT) form, gB730t. As shown previously, gB730t blocks virus entry into cells, suggesting that gB730t competes with virion gB for a cell receptor. All four mutant proteins retained this function, implying that fusion loop activity is separate from gB-receptor binding. However, unlike WT gB730t, the mutant proteins displayed reduced binding to cells and were either impaired or unable to bind naked, cholesterol-enriched liposomes, suggesting that it may be gB-lipid binding that is disrupted by the mutations. Furthermore, monoclonal antibodies with epitopes proximal to the fusion loops abrogated gB-liposome binding. Taken together, our data suggest that gB associates with lipid membranes via a fusion domain of key hydrophobic and hydrophilic residues and that this domain associates with lipid membranes during fusion.
Figures




Similar articles
-
The membrane-proximal region (MPR) of herpes simplex virus gB regulates association of the fusion loops with lipid membranes.mBio. 2012 Nov 20;3(6):e00429-12. doi: 10.1128/mBio.00429-12. mBio. 2012. PMID: 23170000 Free PMC article.
-
Multiple Sites on Glycoprotein H (gH) Functionally Interact with the gB Fusion Protein to Promote Fusion during Herpes Simplex Virus (HSV) Entry.mBio. 2023 Feb 28;14(1):e0336822. doi: 10.1128/mbio.03368-22. Epub 2023 Jan 11. mBio. 2023. PMID: 36629412 Free PMC article.
-
Regulation of herpes simplex virus gB-induced cell-cell fusion by mutant forms of gH/gL in the absence of gD and cellular receptors.mBio. 2013 Feb 26;4(2):e00046-13. doi: 10.1128/mBio.00046-13. mBio. 2013. PMID: 23443004 Free PMC article.
-
Two Sides to Every Story: Herpes Simplex Type-1 Viral Glycoproteins gB, gD, gH/gL, gK, and Cellular Receptors Function as Key Players in Membrane Fusion.Viruses. 2021 Sep 16;13(9):1849. doi: 10.3390/v13091849. Viruses. 2021. PMID: 34578430 Free PMC article. Review.
-
The structural basis of herpesvirus entry.Nat Rev Microbiol. 2021 Feb;19(2):110-121. doi: 10.1038/s41579-020-00448-w. Epub 2020 Oct 21. Nat Rev Microbiol. 2021. PMID: 33087881 Free PMC article. Review.
Cited by
-
Herpes virus fusion and entry: a story with many characters.Viruses. 2012 May;4(5):800-32. doi: 10.3390/v4050800. Epub 2012 May 10. Viruses. 2012. PMID: 22754650 Free PMC article. Review.
-
Low pH-induced conformational change in herpes simplex virus glycoprotein B.J Virol. 2010 Apr;84(8):3759-66. doi: 10.1128/JVI.02573-09. Epub 2010 Feb 10. J Virol. 2010. PMID: 20147407 Free PMC article.
-
Epstein-Barr virus glycoprotein gB and gHgL can mediate fusion and entry in trans, and heat can act as a partial surrogate for gHgL and trigger a conformational change in gB.J Virol. 2014 Nov;88(21):12193-201. doi: 10.1128/JVI.01597-14. Epub 2014 Aug 20. J Virol. 2014. PMID: 25142593 Free PMC article.
-
Human cytomegalovirus (HCMV) glycoprotein gB promotes virus entry in trans acting as the viral fusion protein rather than as a receptor-binding protein.mBio. 2013 Jun 4;4(3):e00332-13. doi: 10.1128/mBio.00332-13. mBio. 2013. PMID: 23736286 Free PMC article.
-
Functional analysis of the Autographa californica multiple nucleopolyhedrovirus GP64 terminal fusion loops and interactions with membranes.J Virol. 2012 Sep;86(18):9617-28. doi: 10.1128/JVI.00813-12. Epub 2012 Jun 27. J Virol. 2012. PMID: 22740400 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

