Disulfide bond formation at the C termini of vaccinia virus A26 and A27 proteins does not require viral redox enzymes and suppresses glycosaminoglycan-mediated cell fusion
- PMID: 19369327
- PMCID: PMC2698572
- DOI: 10.1128/JVI.02295-08
Disulfide bond formation at the C termini of vaccinia virus A26 and A27 proteins does not require viral redox enzymes and suppresses glycosaminoglycan-mediated cell fusion
Abstract
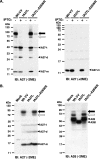
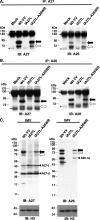
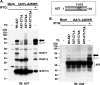
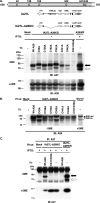
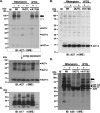
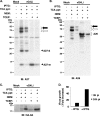
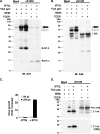
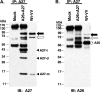
Vaccinia virus A26 protein is an envelope protein of the intracellular mature virus (IMV) of vaccinia virus. A mutant A26 protein with a truncation of the 74 C-terminal amino acids was expressed in infected cells but failed to be incorporated into IMV (W. L. Chiu, C. L. Lin, M. H. Yang, D. L. Tzou, and W. Chang, J. Virol 81:2149-2157, 2007). Here, we demonstrate that A27 protein formed a protein complex with the full-length form but not with the truncated form of A26 protein in infected cells as well as in IMV. The formation of the A26-A27 protein complex occurred prior to virion assembly and did not require another A27-binding protein, A17 protein, in the infected cells. A26 protein contains six cysteine residues, and in vitro mutagenesis showed that Cys441 and Cys442 mediated intermolecular disulfide bonds with Cys71 and Cys72 of viral A27 protein, whereas Cys43 and Cys342 mediated intramolecular disulfide bonds. A26 and A27 proteins formed disulfide-linked complexes in transfected 293T cells, showing that the intermolecular disulfide bond formation did not depend on viral redox pathways. Finally, using cell fusion from within and fusion from without, we demonstrate that cell surface glycosaminoglycan is important for virus-cell fusion and that A26 protein, by forming complexes with A27 protein, partially suppresses fusion.
Figures

Similar articles
-
Crystal structure of vaccinia viral A27 protein reveals a novel structure critical for its function and complex formation with A26 protein.PLoS Pathog. 2013;9(8):e1003563. doi: 10.1371/journal.ppat.1003563. Epub 2013 Aug 22. PLoS Pathog. 2013. PMID: 23990784 Free PMC article.
-
Vaccinia mature virus fusion regulator A26 protein binds to A16 and G9 proteins of the viral entry fusion complex and dissociates from mature virions at low pH.J Virol. 2012 Apr;86(7):3809-18. doi: 10.1128/JVI.06081-11. Epub 2012 Jan 25. J Virol. 2012. PMID: 22278246 Free PMC article.
-
Vaccinia virus A26 and A27 proteins form a stable complex tethered to mature virions by association with the A17 transmembrane protein.J Virol. 2008 Dec;82(24):12384-91. doi: 10.1128/JVI.01524-08. Epub 2008 Oct 8. J Virol. 2008. PMID: 18842719 Free PMC article.
-
Vaccinia viral protein A27 is anchored to the viral membrane via a cooperative interaction with viral membrane protein A17.J Biol Chem. 2014 Mar 7;289(10):6639-6655. doi: 10.1074/jbc.M114.547372. Epub 2014 Jan 22. J Biol Chem. 2014. PMID: 24451374 Free PMC article.
-
Membrane cell fusion activity of the vaccinia virus A17-A27 protein complex.Cell Microbiol. 2008 Jan;10(1):149-64. doi: 10.1111/j.1462-5822.2007.01026.x. Epub 2007 Aug 15. Cell Microbiol. 2008. PMID: 17708756
Cited by
-
Engineered Promoter-Switched Viruses Reveal the Role of Poxvirus Maturation Protein A26 as a Negative Regulator of Viral Spread.J Virol. 2021 Sep 9;95(19):e0101221. doi: 10.1128/JVI.01012-21. Epub 2021 Sep 9. J Virol. 2021. PMID: 34260287 Free PMC article.
-
Smallpox vaccines: targets of protective immunity.Immunol Rev. 2011 Jan;239(1):8-26. doi: 10.1111/j.1600-065X.2010.00975.x. Immunol Rev. 2011. PMID: 21198662 Free PMC article. Review.
-
Elimination of A-type inclusion formation enhances cowpox virus replication in mice: implications for orthopoxvirus evolution.Virology. 2014 Mar;452-453:59-66. doi: 10.1016/j.virol.2013.12.030. Epub 2014 Jan 29. Virology. 2014. PMID: 24606683 Free PMC article.
-
Orthopoxvirus species and strain differences in cell entry.Virology. 2012 Nov 25;433(2):506-12. doi: 10.1016/j.virol.2012.08.044. Epub 2012 Sep 20. Virology. 2012. PMID: 22999097 Free PMC article.
-
Formation of orthopoxvirus cytoplasmic A-type inclusion bodies and embedding of virions are dynamic processes requiring microtubules.J Virol. 2012 May;86(10):5905-14. doi: 10.1128/JVI.06997-11. Epub 2012 Mar 21. J Virol. 2012. PMID: 22438543 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

