Many-core algorithms for statistical phylogenetics
- PMID: 19369496
- PMCID: PMC2682525
- DOI: 10.1093/bioinformatics/btp244
Many-core algorithms for statistical phylogenetics
Abstract
Motivation: Statistical phylogenetics is computationally intensive, resulting in considerable attention meted on techniques for parallelization. Codon-based models allow for independent rates of synonymous and replacement substitutions and have the potential to more adequately model the process of protein-coding sequence evolution with a resulting increase in phylogenetic accuracy. Unfortunately, due to the high number of codon states, computational burden has largely thwarted phylogenetic reconstruction under codon models, particularly at the genomic-scale. Here, we describe novel algorithms and methods for evaluating phylogenies under arbitrary molecular evolutionary models on graphics processing units (GPUs), making use of the large number of processing cores to efficiently parallelize calculations even for large state-size models.
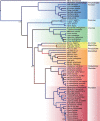
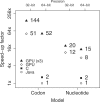
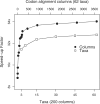
Results: We implement the approach in an existing Bayesian framework and apply the algorithms to estimating the phylogeny of 62 complete mitochondrial genomes of carnivores under a 60-state codon model. We see a near 90-fold speed increase over an optimized CPU-based computation and a >140-fold increase over the currently available implementation, making this the first practical use of codon models for phylogenetic inference over whole mitochondrial or microorganism genomes.
Availability and implementation: Source code provided in BEAGLE: Broad-platform Evolutionary Analysis General Likelihood Evaluator, a cross-platform/processor library for phylogenetic likelihood computation (http://beagle-lib.googlecode.com/). We employ a BEAGLE-implementation using the Bayesian phylogenetics framework BEAST (http://beast.bio.ed.ac.uk/).
Figures

References
-
- Altekar G, et al. Parallel metropolis coupled Markov chain Monte Carlo for Bayesian phylogenetic inference. Bioinformatics. 2004;20:407–415. - PubMed
-
- Bininda-Emonds O, et al. Building large trees by combining phylogenetic information: a complete phylogeny of the extant Carnivora (Mammalia) Biol. Rev. 1999;74:143–175. - PubMed
-
- Charalambous M, et al. Initial experiences porting a bioinformatics application to a graphics processor. In: Bozanis P, Houstis EN, editors. Proceedings of 10th Panhellenic Conference on Informatics (PCI2005). Vol. 3746. New York: Springer; 2005. pp. 415–425. Lecture Notes in Computer Science.
-
- Choi J, et al. PUMMA: parallel universal matrix multiplication algorithms on distributed memory concurrent computers. Concurrency Pract. Exp. 1994;6:543–570.
-
- Delisle I, Strobeck C. A phylogeny of the Caniformia (order Carnivora) based on 12 complete protein-coding mitochondrial genes. Mol. Phylogenet. Evol. 2005;37:192–201. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

