A comparative study of the involvement of 17 Arabidopsis myosin family members on the motility of Golgi and other organelles
- PMID: 19369591
- PMCID: PMC2689979
- DOI: 10.1104/pp.109.136853
A comparative study of the involvement of 17 Arabidopsis myosin family members on the motility of Golgi and other organelles
Abstract
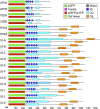
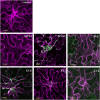
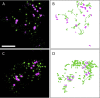
Gene families with multiple members are predicted to have individuals with overlapping functions. We examined all of the Arabidopsis (Arabidopsis thaliana) myosin family members for their involvement in Golgi and other organelle motility. Truncated fragments of all 17 annotated Arabidopsis myosins containing either the IQ tail or tail domains only were fused to fluorescent markers and coexpressed with a Golgi marker in two different plants. We tracked and calculated Golgi body displacement rate in the presence of all myosin truncations and found that tail fragments of myosins MYA1, MYA2, XI-C, XI-E, XI-I, and XI-K were the best inhibitors of Golgi body movement in the two plants. Tail fragments of myosins XI-B, XI-F, XI-H, and ATM1 had an inhibitory effect on Golgi bodies only in Nicotiana tabacum, while tail fragments of myosins XI-G and ATM2 had a slight effect on Golgi body motility only in Nicotiana benthamiana. The best myosin inhibitors of Golgi body motility were able to arrest mitochondrial movement too. No exclusive colocalization was found between these myosins and Golgi bodies in our system, although the excess of cytosolic signal observed could mask myosin molecules bound to the surface of the organelle. From the preserved actin filaments found in the presence of enhanced green fluorescent protein fusions of truncated myosins and the motility of myosin punctae, we conclude that global arrest of actomyosin-derived cytoplasmic streaming had not occurred. Taken together, our data suggest that the above myosins are involved, directly or indirectly, in the movement of Golgi and mitochondria in plant cells.
Figures




References
-
- Baluska F, Samaj J, Hlavacka A, Kendrick-Jones J, Volkmann D (2004) Actin-dependent fluid-phase endocytosis in inner cortex cells of maize root apices. J Exp Bot 55 463–473 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

