Design of protein-interaction specificity gives selective bZIP-binding peptides
- PMID: 19370028
- PMCID: PMC2748673
- DOI: 10.1038/nature07885
Design of protein-interaction specificity gives selective bZIP-binding peptides
Abstract
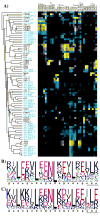
Interaction specificity is a required feature of biological networks and a necessary characteristic of protein or small-molecule reagents and therapeutics. The ability to alter or inhibit protein interactions selectively would advance basic and applied molecular science. Assessing or modelling interaction specificity requires treating multiple competing complexes, which presents computational and experimental challenges. Here we present a computational framework for designing protein-interaction specificity and use it to identify specific peptide partners for human basic-region leucine zipper (bZIP) transcription factors. Protein microarrays were used to characterize designed, synthetic ligands for all but one of 20 bZIP families. The bZIP proteins share strong sequence and structural similarities and thus are challenging targets to bind specifically. Nevertheless, many of the designs, including examples that bind the oncoproteins c-Jun, c-Fos and c-Maf (also called JUN, FOS and MAF, respectively), were selective for their targets over all 19 other families. Collectively, the designs exhibit a wide range of interaction profiles and demonstrate that human bZIPs have only sparsely sampled the possible interaction space accessible to them. Our computational method provides a way to systematically analyse trade-offs between stability and specificity and is suitable for use with many types of structure-scoring functions; thus, it may prove broadly useful as a tool for protein design.
Figures


References
-
- Jones RB, Gordus A, Krall JA, MacBeath G. A quantitative protein interaction network for the ErbB receptors using protein microarrays. Nature. 2006;439:168–74. - PubMed
-
- Wiedemann U, et al. Quantification of PDZ domain specificity, prediction of ligand affinity and rational design of super-binding peptides. J Mol Biol. 2004;343:703–18. - PubMed
-
- Newman JR, Keating AE. Comprehensive identification of human bZIP interactions with coiled-coil arrays. Science. 2003;300:2097–101. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

