Crystal structure of the human lymphoid tyrosine phosphatase catalytic domain: insights into redox regulation
- PMID: 19371084
- PMCID: PMC2747803
- DOI: 10.1021/bi900166y
Crystal structure of the human lymphoid tyrosine phosphatase catalytic domain: insights into redox regulation
Abstract
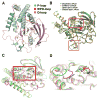
The lymphoid tyrosine phosphatase (LYP), encoded by the PTPN22 gene, recently emerged as an important risk factor and drug target for human autoimmunity. Here we solved the structure of the catalytic domain of LYP, which revealed noticeable differences with previously published structures. The active center with a semi-closed conformation binds a phosphate ion, which may represent an intermediate conformation after dephosphorylation of the substrate but before release of the phosphate product. The structure also revealed an unusual disulfide bond formed between the catalytic Cys and one of the two Cys residues nearby, which is not observed in previously determined structures. Our structural and mutagenesis data suggest that the disulfide bond may play a role in protecting the enzyme from irreversible oxidation. Surprisingly, we found that the two noncatalytic Cys around the active center exert an opposite yin-yang regulation on the catalytic Cys activity. These detailed structural and functional characterizations have provided new insights into autoregulatory mechanisms of LYP function.
Figures

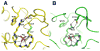

References
-
- Hunter T. A thousand and one protein kinases. Cell. 1987;50:823–829. - PubMed
-
- Mustelin T, Abraham RT, Rudd CE, Alonso A, Merlo JJ. Protein tyrosine phosphorylation in T cell signaling. Frontiers in bioscience : a journal and virtual library. 2002;7:d918–969. - PubMed
-
- Mustelin T, Feng GS, Bottini N, Alonso A, Kholod N, Birle D, Merlo J, Huynh H. Protein tyrosine phosphatases. Frontiers in bioscience : a journal and virtual library. 2002;7:d85–142. - PubMed
-
- Fischer EH, Charbonneau H, Tonks NK. Protein tyrosine phosphatases: a diverse family of intracellular and transmembrane enzymes. Science (New York, NY) 1991;253:401–406. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

