Characterization of the pathogenicity island protein PdpA and its role in the virulence of Francisella novicida
- PMID: 19372153
- PMCID: PMC2889412
- DOI: 10.1099/mic.0.025379-0
Characterization of the pathogenicity island protein PdpA and its role in the virulence of Francisella novicida
Abstract
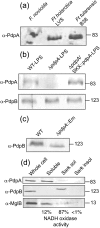
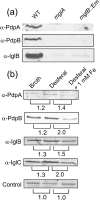
Francisella tularensis is a highly virulent, intracellular pathogen that causes the disease tularaemia. A research surrogate for F. tularensis is Francisella novicida, which causes a tularaemia-like disease in mice, grows similarly in macrophages, and yet is unable to cause disease in humans. Both Francisella species contain a cluster of genes referred to as the Francisella pathogenicity island (FPI). Pathogenicity determinant protein A (PdpA), encoded by the pdpA gene, is located within the FPI and has been associated with the virulence of Francisella species. In this work we examined the properties of PdpA protein expression and localization as well as the phenotype of a F. novicida pdpA deletion mutant. Monoclonal antibody detection of PdpA showed that it is a soluble protein that is upregulated in iron-limiting conditions and undetectable in an mglA or mglB mutant background. Deletion of pdpA resulted in a strain that was highly attenuated for virulence in chicken embryos and mice.
Figures


Similar articles
-
A Francisella novicida pdpA mutant exhibits limited intracellular replication and remains associated with the lysosomal marker LAMP-1.Microbiology (Reading). 2009 May;155(Pt 5):1498-1504. doi: 10.1099/mic.0.025445-0. Epub 2009 Apr 16. Microbiology (Reading). 2009. PMID: 19372155 Free PMC article.
-
Identification of an orphan response regulator required for the virulence of Francisella spp. and transcription of pathogenicity island genes.Infect Immun. 2007 Jul;75(7):3305-14. doi: 10.1128/IAI.00351-07. Epub 2007 Apr 23. Infect Immun. 2007. PMID: 17452468 Free PMC article.
-
Identification of fevR, a novel regulator of virulence gene expression in Francisella novicida.Infect Immun. 2008 Aug;76(8):3473-80. doi: 10.1128/IAI.00430-08. Epub 2008 Jun 16. Infect Immun. 2008. PMID: 18559431 Free PMC article.
-
The Francisella pathogenicity island.Ann N Y Acad Sci. 2007 Jun;1105:122-37. doi: 10.1196/annals.1409.000. Epub 2007 Mar 29. Ann N Y Acad Sci. 2007. PMID: 17395722 Review.
-
Molecular and genetic basis of pathogenesis in Francisella tularensis.Ann N Y Acad Sci. 2007 Jun;1105:138-59. doi: 10.1196/annals.1409.010. Epub 2007 Mar 29. Ann N Y Acad Sci. 2007. PMID: 17395737 Review.
Cited by
-
Disruption of the Francisella noatunensis subsp. orientalis pdpA Gene Results in Virulence Attenuation and Protection in Zebrafish.Infect Immun. 2021 Oct 15;89(11):e0022021. doi: 10.1128/IAI.00220-21. Epub 2021 Aug 23. Infect Immun. 2021. PMID: 34424748 Free PMC article.
-
Working toward the future: insights into Francisella tularensis pathogenesis and vaccine development.Microbiol Mol Biol Rev. 2009 Dec;73(4):684-711. doi: 10.1128/MMBR.00028-09. Microbiol Mol Biol Rev. 2009. PMID: 19946137 Free PMC article. Review.
-
Type A Francisella tularensis acid phosphatases contribute to pathogenesis.PLoS One. 2013;8(2):e56834. doi: 10.1371/journal.pone.0056834. Epub 2013 Feb 15. PLoS One. 2013. PMID: 23457625 Free PMC article.
-
Development, Phenotypic Characterization and Genomic Analysis of a Francisella tularensis Panel for Tularemia Vaccine Testing.Front Microbiol. 2021 Aug 11;12:725776. doi: 10.3389/fmicb.2021.725776. eCollection 2021. Front Microbiol. 2021. PMID: 34456897 Free PMC article.
-
Low dose vaccination with attenuated Francisella tularensis strain SchuS4 mutants protects against tularemia independent of the route of vaccination.PLoS One. 2012;7(5):e37752. doi: 10.1371/journal.pone.0037752. Epub 2012 May 25. PLoS One. 2012. PMID: 22662210 Free PMC article.
References
-
- Angot, A., Peeters, N., Lechner, E., Vailleau, F., Baud, C., Gentzbittel, L., Sartorel, E., Genschik, P., Boucher, C. & Genin, S. (2006). Ralstonia solanacearum requires F-box-like domain-containing type III effectors to promote disease on several host plants. Proc Natl Acad Sci U S A 103, 14620–14625. - PMC - PubMed
-
- Baron, G. S. & Nano, F. E. (1998). MglA and MglB are required for the intramacrophage growth of Francisella novicida. Mol Microbiol 29, 247–259. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

