ALU repeats in promoters are position-dependent co-response elements (coRE) that enhance or repress transcription by dimeric and monomeric progesterone receptors
- PMID: 19372234
- PMCID: PMC2703596
- DOI: 10.1210/me.2009-0048
ALU repeats in promoters are position-dependent co-response elements (coRE) that enhance or repress transcription by dimeric and monomeric progesterone receptors
Abstract
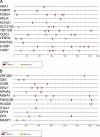
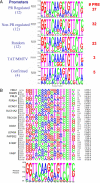
We have conducted an in silico analysis of progesterone response elements (PRE) in progesterone receptor (PR) up-regulated promoters. Imperfect inverted repeats, direct repeats, and half-site PRE are widespread, not only in PR-regulated, but also in non-PR-regulated and random promoters. Few resemble the commonly used palindromic PRE with three nucleotide (nt) spacers. We speculated that PRE may be necessary but insufficient to control endogenous PR-dependent transcription. A search for PRE partners identified a highly conserved 234-nt sequence invariably located within 1-2 kb of transcription start sites. It resembles ALU repeats and contains binding sites for 11 transcription factors. The 234-nt sequence of the PR-regulated 8-oxoguanine DNA glycosylase promoter was cloned in the forward or reverse orientation in front of zero, one, or two inverted repeat PRE, and one or tandem PRE half-sites, driving luciferase. Under these conditions the 234-nt sequence functions as a co-response element (coRE). From the PRE or tandem half-sites, the reverse coRE is a strong activator of PR and glucocorticoid receptor-dependent transcription. The forward coRE is a powerful repressor. The prevalence of PRE half-sites in natural promoters suggested that PR monomers regulate transcription. Indeed, dimerization-domain mutant PR monomers were stronger transactivators than wild-type PR on PRE or tandem half-sites. This was repressed by the forward coRE. We propose that in natural promoters the coRE functions as a composite response element with imperfect PRE and half-sites to present variable, orientation-dependent transcription factors for interaction with nearby PR.
Figures






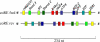
Similar articles
-
The constitution of a progesterone response element.Mol Endocrinol. 1993 Apr;7(4):515-27. doi: 10.1210/mend.7.4.8388996. Mol Endocrinol. 1993. PMID: 8388996
-
Clusters of regulatory signals for RNA polymerase II transcription associated with Alu family repeats and CpG islands in human promoters.Genomics. 2004 May;83(5):873-82. doi: 10.1016/j.ygeno.2003.11.001. Genomics. 2004. PMID: 15081116
-
An Sp1-NF-Y/progesterone receptor DNA binding-dependent mechanism regulates progesterone-induced transcriptional activation of the rabbit RUSH/SMARCA3 gene.J Biol Chem. 2003 Oct 10;278(41):40177-85. doi: 10.1074/jbc.M303921200. Epub 2003 Jul 30. J Biol Chem. 2003. PMID: 12890680
-
Association of some potential hormone response elements in human genes with the Alu family repeats.Gene. 1999 Nov 1;239(2):341-9. doi: 10.1016/s0378-1119(99)00391-1. Gene. 1999. PMID: 10548736
-
Progesterone receptors, their isoforms and progesterone regulated transcription.Mol Cell Endocrinol. 2012 Jun 24;357(1-2):18-29. doi: 10.1016/j.mce.2011.09.016. Epub 2011 Sep 17. Mol Cell Endocrinol. 2012. PMID: 21952082 Free PMC article. Review.
Cited by
-
Control of endothelin-a receptor expression by progesterone is enhanced by synergy with Gata2.Mol Endocrinol. 2013 Jun;27(6):892-908. doi: 10.1210/me.2012-1334. Epub 2013 Apr 16. Mol Endocrinol. 2013. PMID: 23592430 Free PMC article.
-
LINE1-mediated epigenetic repression of androgen receptor transcription causes androgen insensitivity syndrome.Sci Rep. 2024 Jul 15;14(1):16302. doi: 10.1038/s41598-024-65439-w. Sci Rep. 2024. PMID: 39009627 Free PMC article.
-
B2 SINE RNA as a novel regulator of glucocorticoid receptor transcriptional activity.Neurobiol Stress. 2023 Feb 1;23:100522. doi: 10.1016/j.ynstr.2023.100522. eCollection 2023 Mar. Neurobiol Stress. 2023. PMID: 36816533 Free PMC article.
-
Heat shock factor binding in Alu repeats expands its involvement in stress through an antisense mechanism.Genome Biol. 2011 Nov 23;12(11):R117. doi: 10.1186/gb-2011-12-11-r117. Genome Biol. 2011. PMID: 22112862 Free PMC article.
-
SUMOylation Regulates Transcription by the Progesterone Receptor A Isoform in a Target Gene Selective Manner.Diseases. 2018 Jan 2;6(1):5. doi: 10.3390/diseases6010005. Diseases. 2018. PMID: 29301281 Free PMC article.
References
-
- Jantzen HM, Strähle U, Gloss B, Stewart F, Schmid W, Boshart M, Miksicek R, Schütz G 1987 Cooperativity of glucocorticoid response elements located far upstream of the tyrosine aminotransferase gene. Cell 49:29–38 - PubMed
-
- Lieberman BA, Bona BJ, Edwards DP, Nordeen SK 1993 The constitution of a progesterone response element. Mol Endocrinol 7:515–527 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

