Molecular mechanism of poly(ADP-ribosyl)ation by PARP1 and identification of lysine residues as ADP-ribose acceptor sites
- PMID: 19372272
- PMCID: PMC2699514
- DOI: 10.1093/nar/gkp229
Molecular mechanism of poly(ADP-ribosyl)ation by PARP1 and identification of lysine residues as ADP-ribose acceptor sites
Abstract
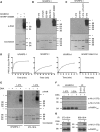
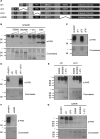
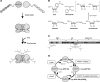
Poly(ADP-ribose) polymerase 1 (PARP1) synthesizes poly(ADP-ribose) (PAR) using nicotinamide adenine dinucleotide (NAD) as a substrate. Despite intensive research on the cellular functions of PARP1, the molecular mechanism of PAR formation has not been comprehensively understood. In this study, we elucidate the molecular mechanisms of poly(ADP-ribosyl)ation and identify PAR acceptor sites. Generation of different chimera proteins revealed that the amino-terminal domains of PARP1, PARP2 and PARP3 cooperate tightly with their corresponding catalytic domains. The DNA-dependent interaction between the amino-terminal DNA-binding domain and the catalytic domain of PARP1 increased V(max) and decreased the K(m) for NAD. Furthermore, we show that glutamic acid residues in the auto-modification domain of PARP1 are not required for PAR formation. Instead, we identify individual lysine residues as acceptor sites for ADP-ribosylation. Together, our findings provide novel mechanistic insights into PAR synthesis with significant relevance for the different biological functions of PARP family members.
Figures




References
-
- Hassa PO, Hottiger MO. The diverse biological roles of mammalian PARPS, a small but powerful family of poly-ADP-ribose polymerases. Front. Biosci. 2008;13:3046–3082. - PubMed
-
- Suzuki H, Uchida K, Shima H, Sato T, Okamoto T, Kimura T, Miwa M. Molecular cloning of cDNA for human poly(ADP-ribose) polymerase and expression of its gene during HL-60 cell differentiation. Biochem. Biophys. Res. Commun. 1987;146:403–409. - PubMed
-
- Kameshita I, Matsuda M, Nishikimi M, Ushiro H, Shizuta Y. Reconstitution and poly(ADP-ribosyl)ation of proteolytically fragmented poly(ADP-ribose) synthetase. J. Biol. Chem. 1986;261:3863–3868. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

