Histone deacetylase Rpd3 antagonizes Sir2-dependent silent chromatin propagation
- PMID: 19372273
- PMCID: PMC2699518
- DOI: 10.1093/nar/gkp233
Histone deacetylase Rpd3 antagonizes Sir2-dependent silent chromatin propagation
Abstract
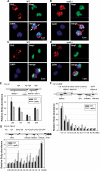
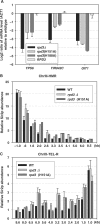
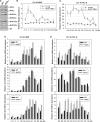
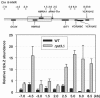
In the eukaryotic genome, transcriptionally silent chromatin tends to propagate along a chromosome and encroach upon adjacent active chromatin. The silencing machinery can be stopped by chromatin boundary elements. We performed a screen in Saccharomyces cerevisiae for proteins that may contribute to the establishment of a chromatin boundary. We found that disruption of histone deacetylase Rpd3p results in defective boundary activity, leading to a Sir-dependent local propagation of transcriptional repression. In rpd3 Delta cells, the amount of Sir2p that was normally found in the nucleolus decreased and the amount of Sir2p found at telomeres and at HM and its adjacent loci increased, leading to an extension of silent chromatin in those areas. In addition, Rpd3p interacted directly with chromatin at boundary regions to deacetylate histone H4 at lysine 5 and at lysine 12. Either the mutation of histone H4 at lysine 5 or a decrease in the histone acetyltransferase (HAT) activity of Esa1p abrogated the silencing phenotype associated with rpd3 mutation, suggesting a novel role for the H4 amino terminus in Rpd3p-mediated heterochromatin boundary regulation. Together, these data provide insight into the molecular mechanisms for the anti-silencing functions of Rpd3p during the formation of heterochromatin boundaries.
Figures



Similar articles
-
Histone Deacetylases with Antagonistic Roles in Saccharomyces cerevisiae Heterochromatin Formation.Genetics. 2016 Sep;204(1):177-90. doi: 10.1534/genetics.116.190835. Epub 2016 Aug 3. Genetics. 2016. PMID: 27489001 Free PMC article.
-
Histone H4 lysine 16 acetylation regulates cellular lifespan.Nature. 2009 Jun 11;459(7248):802-7. doi: 10.1038/nature08085. Nature. 2009. PMID: 19516333 Free PMC article.
-
Rpd3-dependent boundary formation at telomeres by removal of Sir2 substrate.Proc Natl Acad Sci U S A. 2010 Mar 23;107(12):5522-7. doi: 10.1073/pnas.0909169107. Epub 2010 Jan 19. Proc Natl Acad Sci U S A. 2010. PMID: 20133733 Free PMC article.
-
The Nuts and Bolts of Transcriptionally Silent Chromatin in Saccharomyces cerevisiae.Genetics. 2016 Aug;203(4):1563-99. doi: 10.1534/genetics.112.145243. Genetics. 2016. PMID: 27516616 Free PMC article. Review.
-
A model for step-wise assembly of heterochromatin in yeast.Novartis Found Symp. 2004;259:48-56; discussion 56-62, 163-9. Novartis Found Symp. 2004. PMID: 15171246 Review.
Cited by
-
Recruitment of Rpd3 to the telomere depends on the protein arginine methyltransferase Hmt1.PLoS One. 2012;7(8):e44656. doi: 10.1371/journal.pone.0044656. Epub 2012 Aug 31. PLoS One. 2012. PMID: 22953000 Free PMC article.
-
Fkh1 and Fkh2 associate with Sir2 to control CLB2 transcription under normal and oxidative stress conditions.Front Physiol. 2013 Jul 12;4:173. doi: 10.3389/fphys.2013.00173. eCollection 2013. Front Physiol. 2013. PMID: 23874301 Free PMC article.
-
Histone Deacetylases with Antagonistic Roles in Saccharomyces cerevisiae Heterochromatin Formation.Genetics. 2016 Sep;204(1):177-90. doi: 10.1534/genetics.116.190835. Epub 2016 Aug 3. Genetics. 2016. PMID: 27489001 Free PMC article.
-
SWR1 complex poises heterochromatin boundaries for antisilencing activity propagation.Mol Cell Biol. 2010 May;30(10):2391-400. doi: 10.1128/MCB.01106-09. Epub 2010 Mar 22. Mol Cell Biol. 2010. PMID: 20308321 Free PMC article.
-
A sequence-specific interaction between the Saccharomyces cerevisiae rRNA gene repeats and a locus encoding an RNA polymerase I subunit affects ribosomal DNA stability.Mol Cell Biol. 2015 Feb;35(3):544-54. doi: 10.1128/MCB.01249-14. Epub 2014 Nov 24. Mol Cell Biol. 2015. PMID: 25421713 Free PMC article.
References
-
- Richards EJ, Elgin SC. Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects. Cell. 2002;108:489–500. - PubMed
-
- Moazed D. Common themes in mechanisms of gene silencing. Mol. Cell. 2001;8:489–498. - PubMed
-
- Allshire RC, Javerzat JP, Redhead NJ, Cranston G. Position effect variegation at fission yeast centromeres. Cell. 1994;76:157–169. - PubMed
-
- Vermaak D, Ahmad K, Henikoff S. Maintenance of chromatin states: an open-and-shut case. Curr. Opin. Cell Biol. 2003;15:266–274. - PubMed

