Examination of the contributions of size and avidity to the neutralization mechanisms of the anti-HIV antibodies b12 and 4E10
- PMID: 19372381
- PMCID: PMC2678646
- DOI: 10.1073/pnas.0811427106
Examination of the contributions of size and avidity to the neutralization mechanisms of the anti-HIV antibodies b12 and 4E10
Abstract
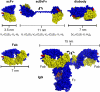
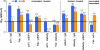
Monoclonal antibodies b12 and 4E10 are broadly neutralizing against a variety of strains of the human immunodeficiency virus type 1 (HIV-1). The epitope for b12 maps to the CD4-binding site in the gp120 subunit of HIV-1's trimeric gp120-gp41 envelope spike, whereas 4E10 recognizes the membrane-proximal external region (MPER) of gp41. Here, we constructed and compared a series of architectures for the b12 and 4E10 combining sites that differed in size, valency, and flexibility. In a comparative analysis of the ability of the b12 and 4E10 constructs to neutralize a panel of clade B HIV-1 strains, we observed that the ability of bivalent constructs to cross-link envelope spikes on the virion surface made a greater contribution to neutralization by b12 than by 4E10. Increased distance and flexibility between antibody combining sites correlated with enhanced neutralization for both antibodies, suggesting restricted mobility for the trimeric spikes embedded in the virion surface. The size of a construct did not appear to be correlated with neutralization potency for b12, but larger 4E10 constructs exhibited a steric occlusion effect, which we interpret as evidence for restricted access to its gp41 epitope. The combination of limited avidity and steric occlusion suggests a mechanism for evading neutralization by antibodies that target epitopes in the highly conserved MPER of gp41.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Wei X, et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. - PubMed
-
- Chen B, et al. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature. 2005;433:834–841. - PubMed
-
- Kwong PD, et al. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature. 2002;420:678–682. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

