X-ray structure of the ternary MTX.NADPH complex of the anthrax dihydrofolate reductase: a pharmacophore for dual-site inhibitor design
- PMID: 19374017
- PMCID: PMC2738603
- DOI: 10.1016/j.jsb.2009.01.001
X-ray structure of the ternary MTX.NADPH complex of the anthrax dihydrofolate reductase: a pharmacophore for dual-site inhibitor design
Abstract
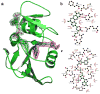
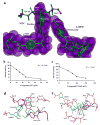
For reasons of bioterrorism and drug resistance, it is imperative to identify and develop new molecular points of intervention against anthrax. Dihydrofolate reductase (DHFR) is a highly conserved enzyme and an established target in a number of species for a variety of chemotherapeutic programs. Recently, the crystal structure of Bacillus anthracis DHFR (baDHFR) in complex with methotrexate (MTX) was determined and, based on the structure, proposals were made for drug design strategies directed against the substrate-binding site. However, little is gleaned about the binding site for NADPH, the cofactor responsible for hydride transfer in the catalytic mechanism. In the present study, X-ray crystallography at 100 K was used to determine the structure of baDHFR in complex with MTX and NADPH. Although the NADPH binding mode is nearly identical to that seen in other DHFR ternary complex structures, the adenine moiety adopts an off-plane tilt of nearly 90 degrees and this orientation is stabilized by hydrogen bonds to functionally conserved Arg residues. A comparison of the binding site, focusing on this region, between baDHFR and the human enzyme is discussed, with an aim at designing species-selective therapeutics. Indeed, the ternary model, refined to 2.3 A resolution, provides an accurate template for testing the feasibility of identifying dual-site inhibitors, compounds that target both the substrate and cofactor-binding site. With the ternary model in hand, using in silico methods, several compounds were identified which could potentially form key bonding contacts in the substrate and cofactor-binding sites. Ultimately, two structurally distinct compounds were verified that inhibit baDHFR at low microM concentrations. The apparent Kd for one of these, (2-(3-(2-(hydroxyimino)-2-(pyridine-4-yl)-6,7-dimethylquinoxalin-2-yl)-1-(pyridine-4-yl)ethanone oxime), was measured by fluorescence spectroscopy to be 5.3 microM.
Figures


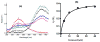
Similar articles
-
Crystal structure of the anthrax drug target, Bacillus anthracis dihydrofolate reductase.J Med Chem. 2007 Sep 6;50(18):4374-81. doi: 10.1021/jm070319v. Epub 2007 Aug 14. J Med Chem. 2007. PMID: 17696333
-
Understanding the role of Leu22 variants in methotrexate resistance: comparison of wild-type and Leu22Arg variant mouse and human dihydrofolate reductase ternary crystal complexes with methotrexate and NADPH.Acta Crystallogr D Biol Crystallogr. 2005 Feb;61(Pt 2):147-55. doi: 10.1107/S0907444904030422. Epub 2005 Jan 19. Acta Crystallogr D Biol Crystallogr. 2005. PMID: 15681865
-
Structure-based enzyme inhibitor design: modeling studies and crystal structure analysis of Pneumocystis carinii dihydrofolate reductase ternary complex with PT653 and NADPH.Acta Crystallogr D Biol Crystallogr. 2002 Jun;58(Pt 6 Pt 2):946-54. doi: 10.1107/s090744490200505x. Epub 2002 May 29. Acta Crystallogr D Biol Crystallogr. 2002. PMID: 12037296
-
The solution structure of Bacillus anthracis dihydrofolate reductase yields insight into the analysis of structure-activity relationships for novel inhibitors.Biochemistry. 2009 May 19;48(19):4100-8. doi: 10.1021/bi802319w. Biochemistry. 2009. PMID: 19323450 Free PMC article.
-
Detection of long-lived bound water molecules in complexes of human dihydrofolate reductase with methotrexate and NADPH.J Mol Biol. 1995 Mar 24;247(2):294-308. doi: 10.1006/jmbi.1994.0140. J Mol Biol. 1995. PMID: 7707376
Cited by
-
Charged Nonclassical Antifolates with Activity Against Gram-Positive and Gram-Negative Pathogens.ACS Med Chem Lett. 2016 May 5;7(7):692-6. doi: 10.1021/acsmedchemlett.6b00120. eCollection 2016 Jul 14. ACS Med Chem Lett. 2016. PMID: 27437079 Free PMC article.
-
Classification of ligand molecules in PDB with graph match-based structural superposition.J Struct Funct Genomics. 2016 Dec;17(4):135-146. doi: 10.1007/s10969-016-9209-x. Epub 2016 Dec 23. J Struct Funct Genomics. 2016. PMID: 28012138
-
Hypothesis-Driven, Structure-Based Design in Photopharmacology: The Case of eDHFR Inhibitors.J Med Chem. 2022 Mar 24;65(6):4798-4817. doi: 10.1021/acs.jmedchem.1c01962. Epub 2022 Mar 8. J Med Chem. 2022. PMID: 35258959 Free PMC article.
-
Solution Ionic Strength Can Modulate Functional Loop Conformations in E. coli Dihydrofolate Reductase.J Phys Chem B. 2024 May 2;128(17):4111-4122. doi: 10.1021/acs.jpcb.4c00677. Epub 2024 Apr 23. J Phys Chem B. 2024. PMID: 38651832 Free PMC article.
-
Advances in Applying Computer-Aided Drug Design for Neurodegenerative Diseases.Int J Mol Sci. 2021 Apr 28;22(9):4688. doi: 10.3390/ijms22094688. Int J Mol Sci. 2021. PMID: 33925236 Free PMC article. Review.
References
-
- Appleman JR, Howell EE, Kraut J, Kuhl M, Blakley RL. Role of aspartate 27 in the binding of methotrexate to dihydrofolate reductase from Escherichia coli. J Biol Chem. 1988;263:9187–98. - PubMed
-
- Argyrou A, Vetting MW, Aladegbami B, Blanchard JS. Mycobacterium tuberculosis dihydrofolate reductase is a target for isoniazid. Nat Struct Mol Biol. 2006;13:408–13. - PubMed
-
- Benkovic SJ. On the mechanism of action of folate- and biopterin-requiring enzymes. Annu Rev Biochem. 1980;49:227–51. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
