Chromatin conformation signatures of cellular differentiation
- PMID: 19374771
- PMCID: PMC2688928
- DOI: 10.1186/gb-2009-10-4-r37
Chromatin conformation signatures of cellular differentiation
Abstract
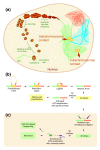
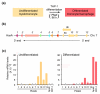
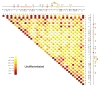
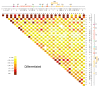
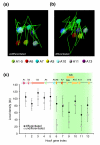
One of the major genomics challenges is to better understand how correct gene expression is orchestrated. Recent studies have shown how spatial chromatin organization is critical in the regulation of gene expression. Here, we developed a suite of computer programs to identify chromatin conformation signatures with 5C technology http://Dostielab.biochem.mcgill.ca. We identified dynamic HoxA cluster chromatin conformation signatures associated with cellular differentiation. Genome-wide chromatin conformation signature identification might uniquely identify disease-associated states and represent an entirely novel class of human disease biomarkers.
Figures

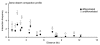

References
-
- Sell S. Leukemia: stem cells, maturation arrest, and differentiation therapy. Stem Cell Rev. 2005;1:197–205. - PubMed
-
- Rosenbauer F, Tenen DG. Transcription factors in myeloid development: balancing differentiation with transformation. Nat Rev Immunol. 2007;7:105–117. - PubMed
-
- The FANTOM Consortium. Suzuki H, Forrest A, van Nimwegen E, Daub C, Balwierz P, Irvine K, Lassman T, Ravasi T, Hasegawa Y, de Hoon M, Katayama S, Schroder K, Carninci P, Akalin A, Ando Y, Arner E, Asada M, Asahara H, Bailey T, Bajic VB, Bauer D, Beckhouse A, Bertin N, Björkegren J, Brombacher F, Bulger E, Chalk AM, Chiba J, Cloonan N, et al. The transcriptional network that controls growth arrest and differentiation in a human myeloid leukemia cell line. Nat Genet. 2009. - PMC - PubMed
-
- West AG, Fraser P. Remote control of gene transcription. Hum Mol Genet. 2005;14:R101–111. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

