Transcriptional features of genomic regulatory blocks
- PMID: 19374772
- PMCID: PMC2688929
- DOI: 10.1186/gb-2009-10-4-r38
Transcriptional features of genomic regulatory blocks
Abstract
Background: Genomic regulatory blocks (GRBs) are chromosomal regions spanned by highly conserved non-coding elements (HCNEs), most of which serve as regulatory inputs of one target gene in the region. The target genes are most often transcription factors involved in embryonic development and differentiation. GRBs often contain extensive gene deserts, as well as additional 'bystander' genes intertwined with HCNEs but whose expression and function are unrelated to those of the target gene. The tight regulation of target genes, complex arrangement of regulatory inputs, and the differential responsiveness of genes in the region call for the examination of fundamental rules governing transcriptional activity in GRBs. Here we use extensive CAGE tag mapping of transcription start sites across different human tissues and differentiation stages combined with expression data and a number of sequence and epigenetic features to discover these rules and patterns.
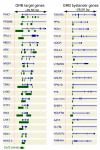
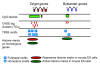
Results: We show evidence that GRB target genes have properties that set them apart from their bystanders as well as other genes in the genome: longer CpG islands, a higher number and wider spacing of alternative transcription start sites, and a distinct composition of transcription factor binding sites in their core/proximal promoters. Target gene expression correlates with the acetylation state of HCNEs in the region. Additionally, target gene promoters have a distinct combination of activating and repressing histone modifications in mouse embryonic stem cell lines.
Conclusions: GRB targets are genes with a number of unique features that are the likely cause of their ability to respond to regulatory inputs from very long distances.
Figures

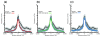




References
-
- Woolfe A, Goodson M, Goode DK, Snell P, McEwen GK, Vavouri T, Smith SF, North P, Callaway H, Kelly K, Walter K, Abnizova I, Gilks W, Edwards YJ, Cooke JE, Elgar G. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 2005;3:e7. doi: 10.1371/journal.pbio.0030007. - DOI - PMC - PubMed
-
- de la Calle-Mustienes E, Feijoo CG, Manzanares M, Tena JJ, Rodriguez-Seguel E, Letizia A, Allende ML, Gomez-Skarmeta JL. A functional survey of the enhancer activity of conserved non-coding sequences from vertebrate Iroquois cluster gene deserts. Genome Res. 2005;15:1061–1072. doi: 10.1101/gr.4004805. - DOI - PMC - PubMed
-
- Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M, Minovitsky S, Dubchak I, Holt A, Lewis KD, Plajzer-Frick I, Akiyama J, De Val S, Afzal V, Black BL, Couronne O, Eisen MB, Visel A, Rubin EM. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. - DOI - PubMed
-
- Kimura-Yoshida C, Kitajima K, Oda-Ishii I, Tian E, Suzuki M, Yamamoto M, Suzuki T, Kobayashi M, Aizawa S, Matsuo I. Characterization of the pufferfish Otx2 cis-regulators reveals evolutionarily conserved genetic mechanisms for vertebrate head specification. Development. 2004;131:57–71. doi: 10.1242/dev.00877. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

