The FANTOM web resource: from mammalian transcriptional landscape to its dynamic regulation
- PMID: 19374775
- PMCID: PMC2688931
- DOI: 10.1186/gb-2009-10-4-r40
The FANTOM web resource: from mammalian transcriptional landscape to its dynamic regulation
Abstract
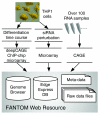
In FANTOM4, an international collaborative research project, we collected a wide range of genome-scale data, including 24 million mRNA 5'-reads (CAGE tags) and microarray expression profiles along a differentiation time course of the human THP-1 cell line and under 52 systematic siRNA perturbations. In addition, data regarding chromatin status derived from ChIP-chip to elucidate the transcriptional regulatory interactions are included. Here we present these data to the research community as an integrated web resource.
Figures


Similar articles
-
Update of the FANTOM web resource: from mammalian transcriptional landscape to its dynamic regulation.Nucleic Acids Res. 2011 Jan;39(Database issue):D856-60. doi: 10.1093/nar/gkq1112. Epub 2010 Nov 12. Nucleic Acids Res. 2011. PMID: 21075797 Free PMC article.
-
CoCo: a web application to display, store and curate ChIP-on-chip data integrated with diverse types of gene expression data.Bioinformatics. 2007 Mar 15;23(6):771-3. doi: 10.1093/bioinformatics/btl641. Epub 2007 Jan 17. Bioinformatics. 2007. PMID: 17234641
-
Cistrome: an integrative platform for transcriptional regulation studies.Genome Biol. 2011 Aug 22;12(8):R83. doi: 10.1186/gb-2011-12-8-r83. Genome Biol. 2011. PMID: 21859476 Free PMC article.
-
[ChIP on chip for transcriptional regulatory network analysis].Tanpakushitsu Kakusan Koso. 2004 Dec;49(17 Suppl):2710-6. Tanpakushitsu Kakusan Koso. 2004. PMID: 15669244 Review. Japanese. No abstract available.
-
[Analysis of the transcriptional regulatory network in the model organisms: yeast and sea urchin].Tanpakushitsu Kakusan Koso. 2004 Dec;49(17 Suppl):2742-50. Tanpakushitsu Kakusan Koso. 2004. PMID: 15669249 Review. Japanese. No abstract available.
Cited by
-
Notch-associated lncRNAs profiling circuiting epigenetic modification in colorectal cancer.Cancer Cell Int. 2022 Oct 13;22(1):316. doi: 10.1186/s12935-022-02736-2. Cancer Cell Int. 2022. PMID: 36229883 Free PMC article. Review.
-
TIPR: transcription initiation pattern recognition on a genome scale.Bioinformatics. 2015 Dec 1;31(23):3725-32. doi: 10.1093/bioinformatics/btv464. Epub 2015 Aug 8. Bioinformatics. 2015. PMID: 26254489 Free PMC article.
-
TransmiR v2.0: an updated transcription factor-microRNA regulation database.Nucleic Acids Res. 2019 Jan 8;47(D1):D253-D258. doi: 10.1093/nar/gky1023. Nucleic Acids Res. 2019. PMID: 30371815 Free PMC article.
-
A large fraction of extragenic RNA pol II transcription sites overlap enhancers.PLoS Biol. 2010 May 11;8(5):e1000384. doi: 10.1371/journal.pbio.1000384. PLoS Biol. 2010. PMID: 20485488 Free PMC article.
-
Update of the FANTOM web resource: from mammalian transcriptional landscape to its dynamic regulation.Nucleic Acids Res. 2011 Jan;39(Database issue):D856-60. doi: 10.1093/nar/gkq1112. Epub 2010 Nov 12. Nucleic Acids Res. 2011. PMID: 21075797 Free PMC article.
References
-
- Okazaki Y, Furuno M, Kasukawa T, Adachi J, Bono H, Kondo S, Nikaido I, Osato N, Saito R, Suzuki H, Yamanaka I, Kiyosawa H, Yagi K, Tomaru Y, Hasegawa Y, Nogami A, Schönbach C, Gojobori T, Baldarelli R, Hill DP, Bult C, Hume DA, Quackenbush J, Schriml LM, Kanapin A, Matsuda H, Batalov S, Beisel KW, Blake JA, Bradt D, et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. doi: 10.1038/nature01266. - DOI - PubMed
-
- Kawai J, Shinagawa A, Shibata K, Yoshino M, Itoh M, Ishii Y, Arakawa T, Hara A, Fukunishi Y, Konno H, Adachi J, Fukuda S, Aizawa K, Izawa M, Nishi K, Kiyosawa H, Kondo S, Yamanaka I, Saito T, Okazaki Y, Gojobori T, Bono H, Kasukawa T, Saito R, Kadota K, Matsuda H, Ashburner M, Batalov S, Casavant T, Fleischmann W, et al. Functional annotation of a full-length mouse cDNA collection. Nature. 2001;409:685–690. doi: 10.1038/35055500. - DOI - PubMed
-
- Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CA, Taylor MS, Engstrom PG, Frith MC, Forrest AR, Alkema WB, Tan SL, Plessy C, Kodzius R, Ravasi T, Kasukawa T, Fukuda S, Kanamori-Katayama M, Kitazume Y, Kawaji H, Kai C, Nakamura M, Konno H, Nakano K, Mottagui-Tabar S, Arner P, Chesi A, Gustincich S, Persichetti F, et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet. 2006;38:626–635. doi: 10.1038/ng1789. - DOI - PubMed
-
- Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, Suzuki H, Carninci P, Hayashizaki Y, Wells C, Frith M, Ravasi T, Pang KC, Hallinan J, Mattick J, Hume DA, Lipovich L, Batalov S, Engström PG, Mizuno Y, Faghihi MA, Sandelin A, Chalk AM, Mottagui-Tabar S, Liang Z, Lenhard B, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. - DOI - PubMed
-
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, Kodzius R, Shimokawa K, Bajic VB, Brenner SE, Batalov S, Forrest AR, Zavolan M, Davis MJ, Wilming LG, Aidinis V, Allen JE, Ambesi-Impiombato A, Apweiler R, Aturaliya RN, Bailey TL, Bansal M, Baxter L, Beisel KW, Bersano T, Bono H, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

