Replisome structure and conformational dynamics underlie fork progression past obstacles
- PMID: 19375905
- PMCID: PMC3732650
- DOI: 10.1016/j.ceb.2009.02.008
Replisome structure and conformational dynamics underlie fork progression past obstacles
Abstract
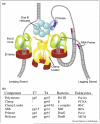
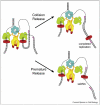
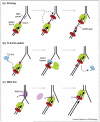
Replisomes are multiprotein complexes that unzip the parental helix and duplicate the separated strands during genome replication. The antiparallel structure of DNA poses unique geometric constraints to the process, and the replisome has evolved unique dynamic features that solve this problem. Interestingly, the solution to duplex DNA replication has been co-opted to solve many other important problems that replisomes must contend with during the duplication of long chromosomes. For example, along its path the replisome will encounter lesions and DNA-bound proteins. Recent studies show that the replisome can circumvent lesions on either strand, using the strategy normally applied to the lagging strand synthesis. Circumventing lesions can also be assisted by other proteins that transiently become a part of the replisome. The replisome must also contend with DNA-binding proteins and recent studies reveal a fascinating process that enables it to bypass RNA polymerase without stopping.
Figures

References
-
- Kornberg A, Baker TA. DNA Replication. edn 2 W.H. Freeman; 1992.
-
- Johnson A, O’Donnell M. Cellular DNA replicases: components and dynamics at the replication fork. Annu Rev Biochem. 2005;74:283–315. - PubMed
-
- Benkovic SJ, Valentine AM, Salinas F. Replisome-mediated DNA replication. Annu Rev Biochem. 2001;70:181–208. - PubMed
-
- McHenry CS. Chromosomal replicases as asymmetric dimers: studies of subunit arrangement and functional consequences. Mol Microbiol. 2003;49:1157–1165. - PubMed
-
- Stillman B. Three-dimensional structure of the beta subunit of E. coli DNA polymerase III holoenzyme: a sliding DNA clamp. Mol Cell. 2008;30:259–260. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

