The role of the retinoblastoma/E2F1 tumor suppressor pathway in the lesion recognition step of nucleotide excision repair
- PMID: 19376752
- PMCID: PMC2700215
- DOI: 10.1016/j.dnarep.2009.03.003
The role of the retinoblastoma/E2F1 tumor suppressor pathway in the lesion recognition step of nucleotide excision repair
Abstract
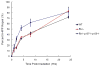
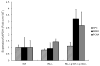
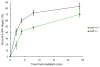
The retinoblastoma Rb/E2F tumor suppressor pathway plays a major role in the regulation of mammalian cell cycle progression. The pRb protein, along with closely related proteins p107 and p130, exerts its anti-proliferative effects by binding to the E2F family of transcription factors known to regulate essential genes throughout the cell cycle. We sought to investigate the role of the Rb/E2F1 pathway in the lesion recognition step of nucleotide excision repair (NER) in mouse embryonic fibroblasts (MEFs). Rb-/-, p107-/-, p130-/- MEFs repaired both cyclobutane pyrimidine dimers (CPDs) and 6-4 photoproducts (6-4PPs) at higher efficiency than did wildtype cells following UV-C irradiation. The expression of damaged DNA binding gene DDB2 involved in the DNA lesion recognition step was elevated in the Rb family-deficient MEFs. To determine if the enhanced DNA repair in the absence of the Rb gene family is due to the derepression of E2F1, we assayed the ability of E2F1-deficient cells to repair damaged DNA and demonstrated that E2F1-/- MEFs are impaired for the removal of both CPDs and 6-4PPs. Furthermore, wildtype cells induced a higher expression of DDB2 and xeroderma pigmentosum gene XPC transcript levels than did E2F1-/- cells following UV-C irradiation. Using an E2F SiteScan algorithm, we uncovered a putative E2F-responsive element in the XPC promoter upstream of the transcription start site. We showed with chromatin immunoprecipitation assays the binding of E2F1 to the XPC promoter in a UV-dependent manner, suggesting that E2F1 is a transcriptional regulator of XPC. Our study identifies a novel E2F1 gene target and further supports the growing body of evidence that the Rb/E2F1 tumor suppressor pathway is involved in the regulation of the DNA lesion recognition step of nucleotide excision repair.
Conflict of interest statement
The authors declare that there is no conflict of interest.
Figures






References
-
- Hanawalt PC, Ford JM, Lloyd DR. Functional characterization of global genomic DNA repair and its implications for cancer. Mutat Res. 2003;544:107–114. - PubMed
-
- van Steeg H, Kraemer KH. Xeroderma pigmentosum and the role of UV-induced DNA damage in skin cancer. Mol Med Today. 1999;5:86–94. - PubMed
-
- Wood RD. Nucleotide excision repair in mammalian cells. J Biol Chem. 1997;272:23465–23468. - PubMed
-
- Wood RD, Mitchell M, Sgouros J, Lindahl T. Human DNA repair genes. Science. 2001;291:1284–1289. - PubMed
-
- Bohr VA, Smith CA, Okumoto DS, Hanawalt PC. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. 1985;40:359–369. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

