Large-scale Arabidopsis phosphoproteome profiling reveals novel chloroplast kinase substrates and phosphorylation networks
- PMID: 19376835
- PMCID: PMC2689975
- DOI: 10.1104/pp.109.138677
Large-scale Arabidopsis phosphoproteome profiling reveals novel chloroplast kinase substrates and phosphorylation networks
Abstract
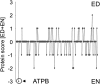
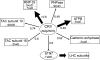
We have characterized the phosphoproteome of Arabidopsis (Arabidopsis thaliana) seedlings using high-accuracy mass spectrometry and report the identification of 1,429 phosphoproteins and 3,029 unique phosphopeptides. Among these, 174 proteins were chloroplast phosphoproteins. Motif-X (motif extractor) analysis of the phosphorylation sites in chloroplast proteins identified four significantly enriched kinase motifs, which include casein kinase II (CKII) and proline-directed kinase motifs, as well as two new motifs at the carboxyl terminus of ribosomal proteins. Using the phosphorylation motifs as a footprint for the activity of a specific kinase class, we connected the phosphoproteins with their putative kinases and constructed a chloroplast CKII phosphorylation network. The network topology suggests that CKII is a central regulator of different chloroplast functions. To provide insights into the dynamic regulation of protein phosphorylation, we analyzed the phosphoproteome at the end of day and end of night. The results revealed only minor changes in chloroplast kinase activities and phosphorylation site utilization. A notable exception was ATP synthase beta-subunit, which is found phosphorylated at CKII phosphorylation sites preferentially in the dark. We propose that ATP synthase is regulated in cooperation with 14-3-3 proteins by CKII-mediated phosphorylation of ATP synthase beta-subunit in the dark.
Figures

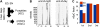

References
-
- Adams JA (2001) Kinetic and catalytic mechanisms of protein kinases. Chem Rev 101 2271–2290 - PubMed
-
- Alexa A, Rahnenfuhrer J, Lengauer T (2006) Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22 1600–1607 - PubMed
-
- Baginsky S (2009) Plant proteomics: concepts, applications, and novel strategies for data interpretation. Mass Spectrom Rev 28 93–120 - PubMed
-
- Baginsky S, Tiller K, Link G (1997) Transcription factor phosphorylation by a protein kinase associated with chloroplast RNA polymerase from mustard (Sinapis alba). Plant Mol Biol 34 181–189 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

