Quorum-sensing control of antibiotic synthesis in Burkholderia thailandensis
- PMID: 19376863
- PMCID: PMC2698390
- DOI: 10.1128/JB.00200-09
Quorum-sensing control of antibiotic synthesis in Burkholderia thailandensis
Abstract
The genome of Burkholderia thailandensis codes for several LuxR-LuxI quorum-sensing systems. We used B. thailandensis quorum-sensing deletion mutants and recombinant Escherichia coli to determine the nature of the signals produced by one of the systems, BtaR2-BtaI2, and to show that this system controls genes required for the synthesis of an antibiotic. BtaI2 is an acyl-homoserine lactone (acyl-HSL) synthase that produces two hydroxylated acyl-HSLs, N-3-hydroxy-decanoyl-HSL (3OHC(10)-HSL) and N-3-hydroxy-octanoyl-HSL (3OHC(8)-HSL). The btaI2 gene is positively regulated by BtaR2 in response to either 3OHC(10)-HSL or 3OHC(8)-HSL. The btaR2-btaI2 genes are located within clusters of genes with annotations that suggest they are involved in the synthesis of polyketide or peptide antibiotics. Stationary-phase cultures of wild-type B. thailandensis, but not a btaR2 mutant or a strain deficient in acyl-HSL synthesis, produced an antibiotic effective against gram-positive bacteria. Two of the putative antibiotic synthesis gene clusters require BtaR2 and either 3OHC(10)-HSL or 3OHC(8)-HSL for activation. This represents another example where antibiotic synthesis is controlled by quorum sensing, and it has implications for the evolutionary divergence of B. thailandensis and its close relatives Burkholderia pseudomallei and Burkholderia mallei.
Figures
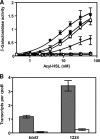




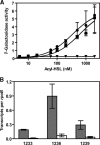
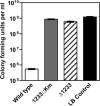
References
-
- Bassler, B. L. 2002. Small talk. Cell-to-cell communication in bacteria. Cell 109421-424. - PubMed
-
- Brett, P. J., D. DeShazer, and D. E. Woods. 1998. Burkholderia thailandensis sp. nov., a Burkholderia pseudomallei-like species. Int. J. Syst. Bacteriol. 48317-320. - PubMed
-
- Burkholder, P. R., and N. H. Giles. 1947. Induced biochemical mutations in Bacillus subtilis. Am. J. Bot. 34345-348. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

