Transcriptomic analysis of Rhizobium leguminosarum biovar viciae in symbiosis with host plants Pisum sativum and Vicia cracca
- PMID: 19376875
- PMCID: PMC2698398
- DOI: 10.1128/JB.00165-09
Transcriptomic analysis of Rhizobium leguminosarum biovar viciae in symbiosis with host plants Pisum sativum and Vicia cracca
Abstract
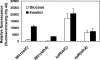
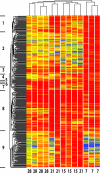
Rhizobium leguminosarum bv. viciae forms nitrogen-fixing nodules on several legumes, including pea (Pisum sativum) and vetch (Vicia cracca), and has been widely used as a model to study nodule biochemistry. To understand the complex biochemical and developmental changes undergone by R. leguminosarum bv. viciae during bacteroid development, microarray experiments were first performed with cultured bacteria grown on a variety of carbon substrates (glucose, pyruvate, succinate, inositol, acetate, and acetoacetate) and then compared to bacteroids. Bacteroid metabolism is essentially that of dicarboxylate-grown cells (i.e., induction of dicarboxylate transport, gluconeogenesis and alanine synthesis, and repression of sugar utilization). The decarboxylating arm of the tricarboxylic acid cycle is highly induced, as is gamma-aminobutyrate metabolism, particularly in bacteroids from early (7-day) nodules. To investigate bacteroid development, gene expression in bacteroids was analyzed at 7, 15, and 21 days postinoculation of peas. This revealed that bacterial rRNA isolated from pea, but not vetch, is extensively processed in mature bacteroids. In early development (7 days), there were large changes in the expression of regulators, exported and cell surface molecules, multidrug exporters, and heat and cold shock proteins. fix genes were induced early but continued to increase in mature bacteroids, while nif genes were induced strongly in older bacteroids. Mutation of 37 genes that were strongly upregulated in mature bacteroids revealed that none were essential for nitrogen fixation. However, screening of 3,072 mini-Tn5 mutants on peas revealed previously uncharacterized genes essential for nitrogen fixation. These encoded a potential magnesium transporter, an AAA domain protein, and proteins involved in cytochrome synthesis.
Figures


Similar articles
-
Transcriptomic analysis of Rhizobium leguminosarum bacteroids in determinate and indeterminate nodules.Microb Genom. 2019 Feb;5(2):e000254. doi: 10.1099/mgen.0.000254. Epub 2019 Feb 19. Microb Genom. 2019. PMID: 30777812 Free PMC article.
-
The hypBFCDE operon from Rhizobium leguminosarum biovar viciae is expressed from an Fnr-type promoter that escapes mutagenesis of the fnrN gene.J Bacteriol. 1995 Oct;177(19):5661-9. doi: 10.1128/jb.177.19.5661-5669.1995. J Bacteriol. 1995. PMID: 7559356 Free PMC article.
-
A microaerobically induced small heat shock protein contributes to Rhizobium leguminosarum/Pisum sativum symbiosis and interacts with a wide range of bacteroid proteins.Appl Environ Microbiol. 2025 Jan 31;91(1):e0138524. doi: 10.1128/aem.01385-24. Epub 2024 Dec 23. Appl Environ Microbiol. 2025. PMID: 39714151 Free PMC article.
-
Legumes regulate Rhizobium bacteroid development and persistence by the supply of branched-chain amino acids.Proc Natl Acad Sci U S A. 2009 Jul 28;106(30):12477-82. doi: 10.1073/pnas.0903653106. Epub 2009 Jul 13. Proc Natl Acad Sci U S A. 2009. PMID: 19597156 Free PMC article.
-
Proteome Analysis Reveals a Significant Host-Specific Response in Rhizobium leguminosarum bv. viciae Endosymbiotic Cells.Mol Cell Proteomics. 2021;20:100009. doi: 10.1074/mcp.RA120.002276. Epub 2020 Dec 6. Mol Cell Proteomics. 2021. PMID: 33214187 Free PMC article.
Cited by
-
Pyruvate is synthesized by two pathways in pea bacteroids with different efficiencies for nitrogen fixation.J Bacteriol. 2010 Oct;192(19):4944-53. doi: 10.1128/JB.00294-10. Epub 2010 Jul 30. J Bacteriol. 2010. PMID: 20675477 Free PMC article.
-
Bacterial Biosensors for in Vivo Spatiotemporal Mapping of Root Secretion.Plant Physiol. 2017 Jul;174(3):1289-1306. doi: 10.1104/pp.16.01302. Epub 2017 May 11. Plant Physiol. 2017. PMID: 28495892 Free PMC article.
-
ABC transport is inactivated by the PTS(Ntr) under potassium limitation in Rhizobium leguminosarum 3841.PLoS One. 2013 May 28;8(5):e64682. doi: 10.1371/journal.pone.0064682. Print 2013. PLoS One. 2013. PMID: 23724079 Free PMC article.
-
The Small-Molecule Language of Dynamic Microbial Interactions.Annu Rev Microbiol. 2022 Sep 8;76:641-660. doi: 10.1146/annurev-micro-042722-091052. Epub 2022 Jun 9. Annu Rev Microbiol. 2022. PMID: 35679616 Free PMC article. Review.
-
Functional Genomics Approaches to Studying Symbioses between Legumes and Nitrogen-Fixing Rhizobia.High Throughput. 2018 May 18;7(2):15. doi: 10.3390/ht7020015. High Throughput. 2018. PMID: 29783718 Free PMC article. Review.
References
-
- Allaway, D., E. Lodwig, L. A. Crompton, M. Wood, T. R. Parsons, T. Wheeler, and P. S. Poole. 2000. Identification of alanine dehydrogenase and its role in mixed secretion of ammonium and alanine by pea bacteroids. Mol. Microbiol. 36508-515. - PubMed
-
- Bahar, M., J. deMajnik, M. Wexler, J. Fry, P. S. Poole, and P. J. Murphy. 1998. A model for the catabolism of rhizopine in Rhizobium leguminosarum involves a ferredoxin oxygenase complex and the inositol degradative pathway. Mol. Plant-Microbe Interact. 111057-1068. - PubMed
-
- Becker, A., H. Berges, E. Krol, C. Bruand, S. Ruberg, D. Capela, E. Lauber, E. Meilhoc, F. Ampe, F. J. de Bruijn, J. Fourment, A. Francez-Charlot, D. Kahn, H. Kuster, C. Liebe, A. Puhler, S. Weidner, and J. Batut. 2004. Global changes in gene expression in Sinorhizobium meliloti 1021 under microoxic and symbiotic conditions. Mol. Plant-Microbe Interact. 17292-303. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

