ATP-binding site lesions in FtsE impair cell division
- PMID: 19376877
- PMCID: PMC2698383
- DOI: 10.1128/JB.00179-09
ATP-binding site lesions in FtsE impair cell division
Abstract
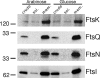
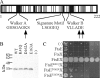
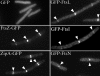
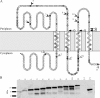
FtsE and FtsX of Escherichia coli constitute an apparent ABC transporter that localizes to the septal ring. In the absence of FtsEX, cells divide poorly and several membrane proteins essential for cell division are largely absent from the septal ring, including FtsK, FtsQ, FtsI, and FtsN. These observations, together with the fact that ftsE and ftsX are cotranscribed with ftsY, which helps to target some proteins for insertion into the cytoplasmic membrane, suggested that FtsEX might contribute to insertion of division proteins into the membrane. Here we show that this hypothesis is probably wrong, because cells depleted of FtsEX had normal amounts of FtsK, FtsQ, FtsI, and FtsN in the membrane fraction. We also show that FtsX localizes to septal rings in cells that lack FtsE, arguing that FtsX targets the FtsEX complex to the ring. Nevertheless, both proteins had to be present to recruit further Fts proteins to the ring. Mutant FtsE proteins with lesions in the ATP-binding site supported septal ring assembly (when produced together with FtsX), but these rings constricted poorly. This finding implies that FtsEX uses ATP to facilitate constriction rather than assembly of the septal ring. Finally, topology analysis revealed that FtsX has only four transmembrane segments, none of which contains a charged amino acid. This structure is not what one would expect of a substrate-specific transmembrane channel, leading us to suggest that FtsEX is not really a transporter even though it probably has to hydrolyze ATP to support cell division.
Figures




References
-
- Aarsman, M. E., A. Piette, C. Fraipont, T. M. Vinkenvleugel, M. Nguyen-Disteche, and T. den Blaauwen. 2005. Maturation of the Escherichia coli divisome occurs in two steps. Mol. Microbiol. 551631-1645. - PubMed
-
- Addinall, S. G., C. Cao, and J. Lutkenhaus. 1997. FtsN, a late recruit to the septum in Escherichia coli. Mol. Microbiol. 25303-309. - PubMed
-
- Arends, S. J. R., K. B. Williams, R. J. Kustusch, and D. S. Weiss. 2007. Cell division, p. 173-197. In M. Ehrmann (ed.), The periplasm. ASM Press, Washington, DC.
-
- Ausubel, F. A., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1998. Current protocols in molecular biology. John Wiley & Sons, New York, NY.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

