Microarray identification of Clostridium difficile core components and divergent regions associated with host origin
- PMID: 19376880
- PMCID: PMC2698405
- DOI: 10.1128/JB.00222-09
Microarray identification of Clostridium difficile core components and divergent regions associated with host origin
Abstract
Clostridium difficile is a gram-positive, spore-forming enteric anaerobe which can infect humans and a wide variety of animal species. Recently, the incidence and severity of human C. difficile infection has markedly increased. In this study, we evaluated the genomic content of 73 C. difficile strains isolated from humans, horses, cattle, and pigs by comparative genomic hybridization with microarrays containing coding sequences from C. difficile strains 630 and QCD-32g58. The sequenced genome of C. difficile strain 630 was used as a reference to define a candidate core genome of C. difficile and to explore correlations between host origins and genetic diversity. Approximately 16% of the genes in strain 630 were highly conserved among all strains, representing the core complement of functional genes defining C. difficile. Absent or divergent genes in the tested strains were distributed across the entire C. difficile 630 genome and across all the predicted functional categories. Interestingly, certain genes were conserved among strains from a specific host species, but divergent in isolates with other host origins. This information provides insight into the genomic changes which might contribute to host adaptation. Due to a high degree of divergence among C. difficile strains, a core gene list from this study offers the first step toward the construction of diagnostic arrays for C. difficile.
Figures



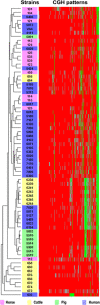
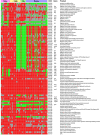

Similar articles
-
Clostridium difficile toxinotype V, ribotype 078, in animals and humans.J Clin Microbiol. 2008 Jun;46(6):2146. doi: 10.1128/JCM.00598-08. Epub 2008 Apr 16. J Clin Microbiol. 2008. PMID: 18417662 Free PMC article. No abstract available.
-
Prevalence of PCR ribotypes among Clostridium difficile isolates from pigs, calves, and other species.J Clin Microbiol. 2007 Jun;45(6):1963-4. doi: 10.1128/JCM.00224-07. Epub 2007 Apr 11. J Clin Microbiol. 2007. PMID: 17428945 Free PMC article.
-
Clostridium difficile Infection in Production Animals and Avian Species: A Review.Foodborne Pathog Dis. 2016 Dec;13(12):647-655. doi: 10.1089/fpd.2016.2181. Epub 2016 Sep 7. Foodborne Pathog Dis. 2016. PMID: 27602596 Review.
-
The Clostridium difficile PCR ribotype 027 lineage: a pathogen on the move.Clin Microbiol Infect. 2014 May;20(5):396-404. doi: 10.1111/1469-0691.12619. Epub 2014 Apr 28. Clin Microbiol Infect. 2014. PMID: 24621128 Review.
-
Array comparative hybridisation reveals a high degree of similarity between UK and European clinical isolates of hypervirulent Clostridium difficile.BMC Genomics. 2010 Jun 21;11:389. doi: 10.1186/1471-2164-11-389. BMC Genomics. 2010. PMID: 20565959 Free PMC article.
Cited by
-
The ecology and pathobiology of Clostridium difficile infections: an interdisciplinary challenge.Zoonoses Public Health. 2011 Feb;58(1):4-20. doi: 10.1111/j.1863-2378.2010.01352.x. Epub 2010 Sep 24. Zoonoses Public Health. 2011. PMID: 21223531 Free PMC article. Review.
-
Comparative transcriptional analysis of clinically relevant heat stress response in Clostridium difficile strain 630.PLoS One. 2012;7(7):e42410. doi: 10.1371/journal.pone.0042410. Epub 2012 Jul 30. PLoS One. 2012. PMID: 22860125 Free PMC article.
-
Diversity and Evolution in the Genome of Clostridium difficile.Clin Microbiol Rev. 2015 Jul;28(3):721-41. doi: 10.1128/CMR.00127-14. Clin Microbiol Rev. 2015. PMID: 26085550 Free PMC article. Review.
-
Genome Analysis of Clostridium difficile PCR Ribotype 014 Lineage in Australian Pigs and Humans Reveals a Diverse Genetic Repertoire and Signatures of Long-Range Interspecies Transmission.Front Microbiol. 2017 Jan 11;7:2138. doi: 10.3389/fmicb.2016.02138. eCollection 2016. Front Microbiol. 2017. PMID: 28123380 Free PMC article.
-
Manganese binds to Clostridium difficile Fbp68 and is essential for fibronectin binding.J Biol Chem. 2011 Feb 4;286(5):3957-69. doi: 10.1074/jbc.M110.184523. Epub 2010 Nov 9. J Biol Chem. 2011. PMID: 21062746 Free PMC article.
References
-
- al-Barrak, A., J. Embil, B. Dyck, K. Olekson, D. Nicoll, M. Alfa, and A. Kabani. 1999. An outbreak of toxin A negative, toxin B positive Clostridium difficile-associated diarrhea in a Canadian tertiary-care hospital. Can. Commun. Dis. Rep. 2565-69. - PubMed
-
- Alfa, M. J., A. Kabani, D. Lyerly, S. Moncrief, L. M. Neville, A. Al-Barrak, G. K. Harding, B. Dyck, K. Olekson, and J. M. Embil. 2000. Characterization of a toxin A-negative, toxin B-positive strain of Clostridium difficile responsible for a nosocomial outbreak of Clostridium difficile-associated diarrhea. J. Clin. Microbiol. 382706-2714. - PMC - PubMed
-
- Allen, M. J., and W. M. Yen (ed.). 2002. Introduction to measurement theory. Waveland Press, Long Grove, IL.
-
- Arroyo, L. G., S. A. Kruth, B. M. Willey, H. R. Staempfli, D. E. Low, and J. S. Weese. 2005. PCR ribotyping of Clostridium difficile isolates originating from human and animal sources. J. Med. Microbiol. 54163-166. - PubMed
-
- Baesman, S. M., T. D. Bullen, J. Dewald, D. Zhang, S. Curran, F. S. Islam, T. J. Beveridge, and R. S. Oremland. 2007. Formation of tellurium nanocrystals during anaerobic growth of bacteria that use Te oxyanions as respiratory electron acceptors. Appl. Environ. Microbiol. 732135-2143. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

