Transcriptome and secretome analyses of Phanerochaete chrysosporium reveal complex patterns of gene expression
- PMID: 19376920
- PMCID: PMC2698378
- DOI: 10.1128/AEM.00314-09
Transcriptome and secretome analyses of Phanerochaete chrysosporium reveal complex patterns of gene expression
Abstract
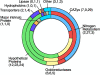
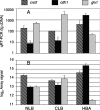
The wood decay basidiomycete Phanerochaete chrysosporium was grown under standard ligninolytic or cellulolytic conditions and subjected to whole-genome expression microarray analysis and liquid chromatography-tandem mass spectrometry of extracellular proteins. A total of 545 genes were flagged on the basis of significant changes in transcript accumulation and/or peptide sequences of the secreted proteins. Under nitrogen or carbon limitation, lignin and manganese peroxidase expression increased relative to nutrient replete medium. Various extracellular oxidases were also secreted in these media, supporting a physiological connection based on peroxide generation. Numerous genes presumed to be involved in mobilizing and recycling nitrogen were expressed under nitrogen limitation, and among these were several secreted glutamic acid proteases not previously observed. In medium containing microcrystalline cellulose as the sole carbon source, numerous genes encoding carbohydrate-active enzymes were upregulated. Among these were six members of the glycoside hydrolase family 61, as well as several polysaccharide lyases and carbohydrate esterases. Presenting a daunting challenge for future research, more than 190 upregulated genes are predicted to encode proteins of unknown function. Of these hypothetical proteins, approximately one-third featured predicted secretion signals, and 54 encoded proteins detected in extracellular filtrates. Our results affirm the importance of certain oxidative enzymes and, underscoring the complexity of lignocellulose degradation, also support an important role for many new proteins of unknown function.
Figures


References
-
- Abbas, A., H. Koc, F. Liu, and M. Tien. 2004. Fungal degradation of wood: initial proteomic analysis of extracellular proteins of Phanerochaete chrysosporium grown on oak substrate. Curr. Genet. 47:49-56. - PubMed
-
- Bao, W., E. Lymar, and V. Renganathan. 1994. Optimization of cellobiose dehydrogenase and β-glucosidase production by cellulose-degrading cultures of Phanerochaete chrysosporium. Appl. Biochem. Biotechnol. 42:642-646.
-
- Benjdia, M., E. Rikirsch, T. Muller, M. Morel, C. Corratge, S. Zimmermann, M. Chalot, W. B. Frommer, and D. Wipf. 2006. Peptide uptake in the ectomycorrhizal fungus Hebeloma cylindrosporum: characterization of two di- and tripeptide transporters (HcPTR2A and B). New Phytol. 170:401-410. - PubMed
-
- Brazma, A., P. Hingamp, J. Quackenbush, G. Sherlock, P. Spellman, C. Stoeckert, J. Aach, W. Ansorge, C. A. Ball, H. C. Causton, T. Gaasterland, P. Glenisson, F. C. Holstege, I. F. Kim, V. Markowitz, J. C. Matese, H. Parkinson, A. Robinson, U. Sarkans, S. Schulze-Kremer, J. Stewart, R. Taylor, J. Vilo, and M. Vingron. 2001. Minimum information about a microarray experiment (MIAME)—toward standards for microarray data. Nat. Genet. 29:365-371. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

