GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis
- PMID: 19376934
- PMCID: PMC2685620
- DOI: 10.1105/tpc.108.065250
GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis
Abstract
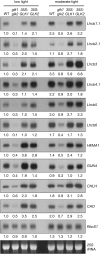
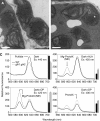
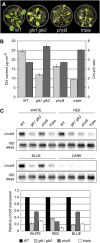
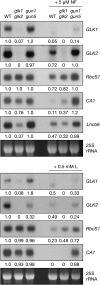
Chloroplasts of photosynthetic organisms harness light energy and convert it into chemical energy. In several land plants, GOLDEN2-LIKE (GLK) transcription factors are required for chloroplast development, as glk1 glk2 double mutants are pale green and deficient in the formation of the photosynthetic apparatus. We show here that glk1 glk2 double mutants of Arabidopsis thaliana accumulate abnormal levels of chlorophyll precursors and that constitutive GLK gene expression leads to increased accumulation of transcripts for antenna proteins and chlorophyll biosynthetic enzymes. To establish the primary targets of GLK gene action, an inducible expression system was used in combination with transcriptome analysis. Following induction, transcript pools were substantially enriched in genes involved in chlorophyll biosynthesis, light harvesting, and electron transport. Chromatin immunoprecipitation experiments confirmed the direct association of GLK1 protein with target gene promoters, revealing a putative regulatory cis-element. We show that GLK proteins influence photosynthetic gene expression independently of the phyB signaling pathway and that the two GLK genes are differentially responsive to plastid retrograde signals. These results suggest that GLK genes help to coregulate and synchronize the expression of a suite of nuclear photosynthetic genes and thus act to optimize photosynthetic capacity in varying environmental and developmental conditions.
Figures






Similar articles
-
Specialization of the Golden2-like regulatory pathway during land plant evolution.New Phytol. 2009;183(1):133-141. doi: 10.1111/j.1469-8137.2009.02829.x. Epub 2009 Mar 30. New Phytol. 2009. PMID: 19383092
-
GLK transcription factors regulate chloroplast development in a cell-autonomous manner.Plant J. 2008 Nov;56(3):432-44. doi: 10.1111/j.1365-313X.2008.03616.x. Epub 2008 Aug 4. Plant J. 2008. PMID: 18643989
-
Photosynthesis of root chloroplasts developed in Arabidopsis lines overexpressing GOLDEN2-LIKE transcription factors.Plant Cell Physiol. 2013 Aug;54(8):1365-77. doi: 10.1093/pcp/pct086. Epub 2013 Jun 7. Plant Cell Physiol. 2013. PMID: 23749810 Free PMC article.
-
Dark-inducible BGH2 suppresses GLK transcription factors and maintains plastid homeostasis to promote light adaptation.Plant Cell. 2025 Aug 4;37(8):koaf180. doi: 10.1093/plcell/koaf180. Plant Cell. 2025. PMID: 40694618
-
Plastid retrograde signaling: A developmental perspective.Plant Cell. 2024 Oct 3;36(10):3903-3913. doi: 10.1093/plcell/koae094. Plant Cell. 2024. PMID: 38546347 Free PMC article. Review.
Cited by
-
A Novel R2R3-MYB Transcription Factor BpMYB106 of Birch (Betula platyphylla) Confers Increased Photosynthesis and Growth Rate through Up-regulating Photosynthetic Gene Expression.Front Plant Sci. 2016 Mar 22;7:315. doi: 10.3389/fpls.2016.00315. eCollection 2016. Front Plant Sci. 2016. PMID: 27047502 Free PMC article.
-
SCARECROW gene function is required for photosynthetic development in maize.Plant Direct. 2020 Sep 9;4(9):e00264. doi: 10.1002/pld3.264. eCollection 2020 Sep. Plant Direct. 2020. PMID: 32999956 Free PMC article.
-
m6A RNA methylation counteracts dark-induced leaf senescence in Arabidopsis.Plant Physiol. 2024 Mar 29;194(4):2663-2678. doi: 10.1093/plphys/kiad660. Plant Physiol. 2024. PMID: 38084897 Free PMC article.
-
The pleiotropic functions of GOLDEN2-LIKE transcription factors in plants.Front Plant Sci. 2024 Aug 19;15:1445875. doi: 10.3389/fpls.2024.1445875. eCollection 2024. Front Plant Sci. 2024. PMID: 39224848 Free PMC article. Review.
-
Coordination of plastid protein import and nuclear gene expression by plastid-to-nucleus retrograde signaling.Plant Physiol. 2009 Nov;151(3):1339-53. doi: 10.1104/pp.109.145987. Epub 2009 Sep 2. Plant Physiol. 2009. PMID: 19726569 Free PMC article.
References
-
- Allen, J.F., and Forsberg, J. (2001). Molecular recognition in thylakoid structure and function. Trends Plant Sci. 6 317–326. - PubMed
-
- Anderson, J.M., Chow, W.S., and Park, Y.-I. (1995). The grand design of photosynthesis: Acclimation of the photosynthetic apparatus to environmental cues. Photosynth. Res. 46 129–139. - PubMed
-
- Aseeva, E., Ossenbühl, F., Sippel, C., Cho, W.K., Stein, B., Eichacker, L.A., Meurer, J., Wanner, G., Westhoff, P., Soll, J., and Vothknecht, U.C. (2007). Vipp1 is required for basic thylakoid membrane formation but not for the assembly of thylakoid protein complexes. Plant Physiol. Biochem. 45 119–128. - PubMed
-
- Balk, J., and Lobrèaux, S. (2005). Biogenesis of iron-sulfur proteins in plants. Trends Plant Sci. 10 324–331. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

