The RNA binding protein ELF9 directly reduces SUPPRESSOR OF OVEREXPRESSION OF CO1 transcript levels in arabidopsis, possibly via nonsense-mediated mRNA decay
- PMID: 19376936
- PMCID: PMC2685614
- DOI: 10.1105/tpc.108.064774
The RNA binding protein ELF9 directly reduces SUPPRESSOR OF OVEREXPRESSION OF CO1 transcript levels in arabidopsis, possibly via nonsense-mediated mRNA decay
Abstract
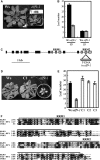
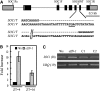
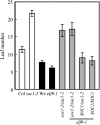
SUPPRESSOR OF OVEREXPRESSION OF CO1 (SOC1) is regulated by a complex transcriptional regulatory network that allows for the integration of multiple floral regulatory inputs from photoperiods, gibberellin, and FLOWERING LOCUS C. However, the posttranscriptional regulation of SOC1 has not been explored. Here, we report that EARLY FLOWERING9 (ELF9), an Arabidopsis thaliana RNA binding protein, directly targets the SOC1 transcript and reduces SOC1 mRNA levels, possibly through a nonsense-mediated mRNA decay (NMD) mechanism, which leads to the degradation of abnormal transcripts with premature translation termination codons (PTCs). The fully spliced SOC1 transcript is upregulated in elf9 mutants as well as in mutants of NMD core components. Furthermore, a partially spliced SOC1 transcript containing a PTC is upregulated more significantly than the fully spliced transcript in elf9 in an ecotype-dependent manner. A Myc-tagged ELF9 protein (MycELF9) directly binds to the partially spliced SOC1 transcript. Previously known NMD target transcripts of Arabidopsis are also upregulated in elf9 and recognized directly by MycELF9. SOC1 transcript levels are also increased by the inhibition of translational activity of the ribosome. Thus, the SOC1 transcript is one of the direct targets of ELF9, which appears to be involved in NMD-dependent mRNA quality control in Arabidopsis.
Figures





References
-
- Abe, M., Kobayashi, Y., Yamamoto, S., Daimon, Y., Yamaguchi, A., Ikeda, Y., Ichinoki, H., Notaguchi, M., Goto, K., and Araki, T. (2005). FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309 1052–1056. - PubMed
-
- Arciga-Reyes, L., Wootton, L., Kieffer, M., and Davies, B. (2006). UPF1 is required for nonsense-mediated mRNA decay (NMD) and RNAi in Arabidopsis. Plant J. 47 480–489. - PubMed
-
- Ausin, I., Alonso-Blanco, C., Jarillo, J.A., Ruiz-Garcia, L., and Martinez-Zapater, J.M. (2004). Regulation of flowering time by FVE, a retinoblastoma-associated protein. Nat. Genet. 36 162–166. - PubMed
-
- Bäurle, I., and Dean, C. (2006). The timing of developmental transitions in plants. Cell 125 655–664. - PubMed
-
- Bezerra, I.C., Michaels, S.D., Schomburg, F.M., and Amasino, R.M. (2004). Lesions in the mRNA cap-binding gene ABA HYPERSENSITIVE 1 suppress FRIGIDA-mediated delayed flowering in Arabidopsis. Plant J. 40 112–119. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

