High resolution mapping of expression QTLs in heterogeneous stock mice in multiple tissues
- PMID: 19376938
- PMCID: PMC2694476
- DOI: 10.1101/gr.088120.108
High resolution mapping of expression QTLs in heterogeneous stock mice in multiple tissues
Abstract
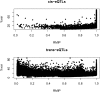
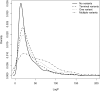
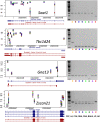
A proportion of the genetic variants underlying complex phenotypes do so through their effects on gene expression, so an important challenge in complex trait analysis is to discover the genetic basis for the variation in transcript abundance. So far, the potential of mapping both quantitative trait loci (QTLs) and expression quantitative trait loci (eQTLs) in rodents has been limited by the low mapping resolution inherent in crosses between inbred strains. We provide a megabase resolution map of thousands of eQTLs in hippocampus, lung, and liver samples from heterogeneous stock (HS) mice in which 843 QTLs have also been mapped at megabase resolution. We exploit dense mouse SNP data to show that artifacts due to allele-specific hybridization occur in approximately 30% of the cis-acting eQTLs and, by comparison with exon expression data, we show that alternative splicing of the 3' end of the genes accounts for <1% of cis-acting eQTLs. Approximately one third of cis-acting eQTLs and one half of trans-acting eQTLs are tissue specific. We have created an important systems biology resource for the genetic analysis of complex traits in a key model organism.
Figures


References
-
- Belknap J.K., Mitchel S.R., Crabbe J.C. Type I and II error rates for quantitative trait loci (QTL) mapping studies using recombinant inbred mouse strains: Computer simulation and empirical results. Behav. Genet. 1996;26:149–160. - PubMed
-
- Bystrykh L., Weersing E., Dontje B., Sutton S., Pletcher M.T., Wiltshire T., Su A.I., Vellenga E., Wang J., Manly K.F., et al. Uncovering regulatory pathways that affect hematopoietic stem cell function using “genetical genomics.”. Nat. Genet. 2005;37:225–232. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
