Seeded growth of beta-amyloid fibrils from Alzheimer's brain-derived fibrils produces a distinct fibril structure
- PMID: 19376973
- PMCID: PMC2678625
- DOI: 10.1073/pnas.0812033106
Seeded growth of beta-amyloid fibrils from Alzheimer's brain-derived fibrils produces a distinct fibril structure
Abstract
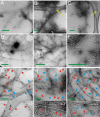
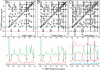
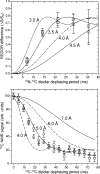
Studies by solid-state nuclear magnetic resonance (NMR) of amyloid fibrils prepared in vitro from synthetic 40-residue beta-amyloid (Abeta(1-40)) peptides have shown that the molecular structure of Abeta(1-40) fibrils is not uniquely determined by amino acid sequence. Instead, the fibril structure depends on the precise details of growth conditions. The molecular structures of beta-amyloid fibrils that develop in Alzheimer's disease (AD) are therefore uncertain. We demonstrate through thioflavin T fluorescence and electron microscopy that fibrils extracted from brain tissue of deceased AD patients can be used to seed the growth of synthetic Abeta(1-40) fibrils, allowing preparation of fibrils with isotopic labeling and in sufficient quantities for solid-state NMR and other measurements. Because amyloid structures propagate themselves in seeded growth, as shown in previous studies, the molecular structures of brain-seeded synthetic Abeta(1-40) fibrils most likely reflect structures that are present in AD brain. Solid-state (13)C NMR spectra of fibril samples seeded with brain material from two AD patients were found to be nearly identical, indicating the same molecular structures. Spectra of an unseeded control sample indicate greater structural heterogeneity. (13)C chemical shifts and other NMR data indicate that the predominant molecular structure in brain-seeded fibrils differs from the structures of purely synthetic Abeta(1-40) fibrils that have been characterized in detail previously. These results demonstrate a new approach to detailed structural characterization of amyloid fibrils that develop in human tissue, and to investigations of possible correlations between fibril structure and the degree of cognitive impairment and neurodegeneration in AD.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Petkova AT, et al. Self-propagating, molecular-level polymorphism in Alzheimer's β-amyloid fibrils. Science. 2005;307:262–265. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

