Diversity-based, model-guided construction of synthetic gene networks with predicted functions
- PMID: 19377462
- PMCID: PMC2680460
- DOI: 10.1038/nbt.1536
Diversity-based, model-guided construction of synthetic gene networks with predicted functions
Abstract
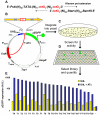
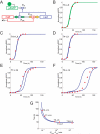
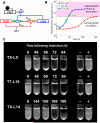
Engineering artificial gene networks from modular components is a major goal of synthetic biology. However, the construction of gene networks with predictable functions remains hampered by a lack of suitable components and the fact that assembled networks often require extensive, iterative retrofitting to work as intended. Here we present an approach that couples libraries of diversified components (synthesized with randomized nonessential sequence) with in silico modeling to guide predictable gene network construction without the need for post hoc tweaking. We demonstrate our approach in Saccharomyces cerevisiae by synthesizing regulatory promoter libraries and using them to construct feed-forward loop networks with different predicted input-output characteristics. We then expand our method to produce a synthetic gene network acting as a predictable timer, modifiable by component choice. We use this network to control the timing of yeast sedimentation, illustrating how the plug-and-play nature of our design can be readily applied to biotechnology.
Figures

Comment in
-
Overpowering the component problem.Nat Biotechnol. 2009 May;27(5):450-1. doi: 10.1038/nbt0509-450. Nat Biotechnol. 2009. PMID: 19430449 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

