Genetic analysis of zinc-finger nuclease-induced gene targeting in Drosophila
- PMID: 19380480
- PMCID: PMC2710147
- DOI: 10.1534/genetics.109.101329
Genetic analysis of zinc-finger nuclease-induced gene targeting in Drosophila
Abstract
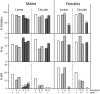
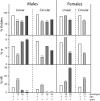
Using zinc-finger nucleases (ZFNs) to cleave the chromosomal target, we have achieved high frequencies of gene targeting in the Drosophila germline. Both local mutagenesis through nonhomologous end joining (NHEJ) and gene replacement via homologous recombination (HR) are stimulated by target cleavage. In this study we investigated the mechanisms that underlie these processes, using materials for the rosy (ry) locus. The frequency of HR dropped significantly in flies homozygous for mutations in spnA (Rad51) or okr (Rad54), two components of the invasion-mediated synthesis-dependent strand annealing (SDSA) pathway. When single-strand annealing (SSA) was also blocked by the use of a circular donor DNA, HR was completely abolished. This indicates that the majority of HR proceeds via SDSA, with a minority mediated by SSA. In flies deficient in lig4 (DNA ligase IV), a component of the major NHEJ pathway, the proportion of HR products rose significantly. This indicates that most NHEJ products are produced in a lig4-dependent process. When both spnA and lig4 were mutated and a circular donor was provided, the frequency of ry mutations was still high and no HR products were recovered. The local mutations produced in these circumstances must have arisen through an alternative, lig4-independent end-joining mechanism. These results show what repair pathways operate on double-strand breaks in this gene targeting system. They also demonstrate that the outcome can be biased toward gene replacement by disabling the major NHEJ pathway and toward simple mutagenesis by interfering with the major HR process.
Figures



References
-
- Adams, M. D., M. McVey and J. J. Sekelsky, 2003. Drosophila BLM in double-strand break repair by synthesis-dependent strand annealing. Science 299 265–267. - PubMed
-
- Barnes, D. E., G. Stamp, I. Rosewell, A. Denzel and T. Lindahl, 1998. Targeted disruption of the gene encoding DNA ligase IV leads to lethality in embryonic mice. Curr. Biol. 8 1395–1398. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

