Single-nucleotide discrimination in immobilized DNA oligonucleotides with a biological nanopore
- PMID: 19380741
- PMCID: PMC2683137
- DOI: 10.1073/pnas.0901054106
Single-nucleotide discrimination in immobilized DNA oligonucleotides with a biological nanopore
Abstract
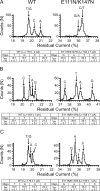
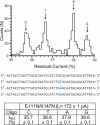
The sequencing of individual DNA strands with nanopores is under investigation as a rapid, low-cost platform in which bases are identified in order as the DNA strand is transported through a pore under an electrical potential. Although the preparation of solid-state nanopores is improving, biological nanopores, such as alpha-hemolysin (alphaHL), are advantageous because they can be precisely manipulated by genetic modification. Here, we show that the transmembrane beta-barrel of an engineered alphaHL pore contains 3 recognition sites that can be used to identify all 4 DNA bases in an immobilized single-stranded DNA molecule, whether they are located in an otherwise homopolymeric DNA strand or in a heteropolymeric strand. The additional steps required to enable nanopore DNA sequencing are outlined.
Conflict of interest statement
Conflict of interest statement: Hagan Bayley is the Founder, a Director, and a shareholder of Oxford Nanopore Technologies, a company engaged in the development of nanopore sequencing technology. This article was not supported by Oxford Nanopore Technologies.
Figures



References
-
- Schloss JA. How to get genomes at one ten-thousandth the cost. Nat Biotechnol. 2008;26:1113–1115. - PubMed
-
- Mardis ER. Next-generation DNA sequencing methods. Annu Rev Genomics Hum Genet. 2008;9:387–402. - PubMed
-
- Bayley H. Sequencing single molecules of DNA. Curr Opin Chem Biol. 2006;10:628–637. - PubMed
-
- Gupta PK. Single-molecule DNA sequencing technologies for future genomics research. Trends Biotechnol. 2008;26:602–611. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

