Transcriptome transfer produces a predictable cellular phenotype
- PMID: 19380745
- PMCID: PMC2670883
- DOI: 10.1073/pnas.0902161106
Transcriptome transfer produces a predictable cellular phenotype
Abstract
Cellular phenotype is the conglomerate of multiple cellular processes involving gene and protein expression that result in the elaboration of a cell's particular morphology and function. It has been thought that differentiated postmitotic cells have their genomes hard wired, with little ability for phenotypic plasticity. Here we show that transfer of the transcriptome from differentiated rat astrocytes into a nondividing differentiated rat neuron resulted in the conversion of the neuron into a functional astrocyte-like cell in a time-dependent manner. This single-cell study permits high resolution of molecular and functional components that underlie phenotype identity. The RNA population from astrocytes contains RNAs in the appropriate relative abundances that give rise to regulatory RNAs and translated proteins that enable astrocyte identity. When transferred into the postmitotic neuron, the astrocyte RNA population converts 44% of the neuronal host cells into the destination astrocyte-like phenotype. In support of this observation, quantitative measures of cellular morphology, single-cell PCR, single-cell microarray, and single-cell functional analyses have been performed. The host-cell phenotypic changes develop over many weeks and are persistent. We call this process of RNA-induced phenotype changes, transcriptome-induced phenotype remodeling.
Conflict of interest statement
Conflict of interest: William T. Greenough and J.E. have collaborated on past research. They are not currently collaborating. The remaining authors declare no conflict of interest.
Figures
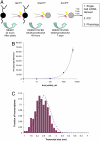

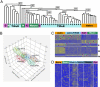

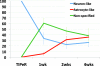
References
-
- Waddington. The Strategy of the Genes: a Discussion of Some Aspects of Theoretical Biology. London: George Allen & Unwin; 1957.
-
- Gurdon JB, Elsdale TR, Fischberg M. Sexually mature individuals of Xenopus laevis from the transplantation of single somatic nuclei. Nature. 1958;182(4627):64–65. - PubMed
-
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. - PubMed
-
- Huangfu D, et al. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat Biotechnol. 2008;26(11):1269–1275. - PubMed
-
- Kim JB, et al. Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors. Nature. 2008;454(7204):646–650. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

