Telomeric RNAs mark sex chromosomes in stem cells
- PMID: 19380904
- PMCID: PMC2710151
- DOI: 10.1534/genetics.109.103093
Telomeric RNAs mark sex chromosomes in stem cells
Abstract
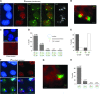
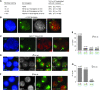
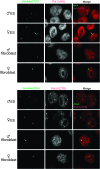
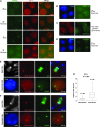
Telomeric regions are known to be transcribed in several organisms. Although originally reported to be transcribed from all chromosomes with enrichment near the inactive X of female cells, we show that telomeric RNAs in fact are enriched on both sex chromosomes of the mouse in a developmentally specific manner. In female stem cells, both active Xs are marked by the RNAs. In male stem cells, both the X and the Y accumulate telomeric RNA. Distribution of telomeric RNAs changes during cell differentiation, after which they associate only with the heterochromatic sex chromosomes of each sex. FISH mapping suggests that accumulated telomeric RNAs localize at the distal telomeric end. Interestingly, telomeric expression changes in cancer and during cellular stress. Furthermore, RNA accumulation increases in Dicer-deficient stem cells, suggesting direct or indirect links to RNAi. We propose that telomeric RNAs are tied to cell differentiation and may be used to mark pluripotency and disease.
Figures




Similar articles
-
X-chromosome inactivation: the molecular basis of silencing.J Biol. 2008 Oct 27;7(8):30. doi: 10.1186/jbiol95. J Biol. 2008. PMID: 18983701 Free PMC article. Review.
-
X chromosome inactivation in the absence of Dicer.Proc Natl Acad Sci U S A. 2009 Jan 27;106(4):1122-7. doi: 10.1073/pnas.0812210106. Epub 2009 Jan 21. Proc Natl Acad Sci U S A. 2009. PMID: 19164542 Free PMC article.
-
Dicer independent small RNAs associate with telomeric heterochromatin.RNA. 2009 Jul;15(7):1274-81. doi: 10.1261/rna.1423309. Epub 2009 May 21. RNA. 2009. PMID: 19460867 Free PMC article.
-
Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends.Science. 2007 Nov 2;318(5851):798-801. doi: 10.1126/science.1147182. Epub 2007 Oct 4. Science. 2007. PMID: 17916692
-
Dicer cuts the kidney.J Am Soc Nephrol. 2008 Nov;19(11):2043-6. doi: 10.1681/ASN.2008090986. Epub 2008 Oct 15. J Am Soc Nephrol. 2008. PMID: 18923053 Review. No abstract available.
Cited by
-
Long Non-Coding RNAs in Neuronal Aging.Noncoding RNA. 2018 Apr 18;4(2):12. doi: 10.3390/ncrna4020012. Noncoding RNA. 2018. PMID: 29670042 Free PMC article. Review.
-
Using amino-labeled nucleotide probes for simultaneous single molecule RNA-DNA FISH.PLoS One. 2014 Sep 16;9(9):e107425. doi: 10.1371/journal.pone.0107425. eCollection 2014. PLoS One. 2014. PMID: 25226542 Free PMC article.
-
Role of TERRA in the regulation of telomere length.Int J Biol Sci. 2015 Feb 5;11(3):316-23. doi: 10.7150/ijbs.10528. eCollection 2015. Int J Biol Sci. 2015. PMID: 25678850 Free PMC article. Review.
-
Aberrant chromatin landscape following loss of the H3.3 chaperone Daxx in haematopoietic precursors leads to Pu.1-mediated neutrophilia and inflammation.Nat Cell Biol. 2021 Dec;23(12):1224-1239. doi: 10.1038/s41556-021-00774-y. Epub 2021 Dec 7. Nat Cell Biol. 2021. PMID: 34876685 Free PMC article.
-
Telomere length regulates TERRA levels through increased trimethylation of telomeric H3K9 and HP1α.Nat Struct Mol Biol. 2012 Sep;19(9):948-56. doi: 10.1038/nsmb.2364. Epub 2012 Aug 26. Nat Struct Mol Biol. 2012. PMID: 22922742
References
-
- Adewumi, O., B. Aflatoonian, L. Ahrlund-Richter, M. Amit, P. W. Andrews et al., 2007. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat. Biotechnol. 25 803–816. - PubMed
-
- Azzalin, C. M., P. Reichenback, L. Khoriauli, E. Giulotto and J. Lingner, 2007. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318 798–801. - PubMed
-
- Bacher, C. P., M. Guggiari, B. Brors, S. Augui, P. Clerc et al., 2006. Transient colocalization of X-inactivation centres accompanies the initiation of X inactivation. Nat. Cell Biol. 8 293–299. - PubMed
-
- Baur, J. A., Y. Zou, J. W. Shay and W. E. Wright, 2001. Telomere position effect in human cells. Science 292 2075–2077. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

