Deletions in the repertoire of Pseudomonas syringae pv. tomato DC3000 type III secretion effector genes reveal functional overlap among effectors
- PMID: 19381254
- PMCID: PMC2663052
- DOI: 10.1371/journal.ppat.1000388
Deletions in the repertoire of Pseudomonas syringae pv. tomato DC3000 type III secretion effector genes reveal functional overlap among effectors
Abstract
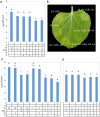
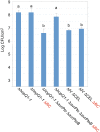
The gamma-proteobacterial plant pathogen Pseudomonas syringae pv. tomato DC3000 uses the type III secretion system to inject ca. 28 Avr/Hop effector proteins into plants, which enables the bacterium to grow from low inoculum levels to produce bacterial speck symptoms in tomato, Arabidopsis thaliana, and (when lacking hopQ1-1) Nicotiana benthamiana. The effectors are collectively essential but individually dispensable for the ability of the bacteria to defeat defenses, grow, and produce symptoms in plants. Eighteen of the effector genes are clustered in six genomic islands/islets. Combinatorial deletions involving these clusters and two of the remaining effector genes revealed a redundancy-based structure in the effector repertoire, such that some deletions diminished growth in N. benthamiana only in combination with other deletions. Much of the ability of DC3000 to grow in N. benthamiana was found to be due to five effectors in two redundant-effector groups (REGs), which appear to separately target two high-level processes in plant defense: perception of external pathogen signals (AvrPto and AvrPtoB) and deployment of antimicrobial factors (AvrE, HopM1, HopR1). Further support for the membership of HopR1 in the same REG as AvrE was gained through bioinformatic analysis, revealing the existence of an AvrE/DspA/E/HopR effector superfamily, which has representatives in virtually all groups of proteobacterial plant pathogens that deploy type III effectors.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Gohre V, Robatzek S. Breaking the barriers: Microbial effector molecules subvert plant immunity. Annu Rev Phytopathol. 2008;46:189–215. - PubMed
-
- Mota LJ, Cornelis GR. The bacterial injection kit: type III secretion systems. Ann Med. 2005;37:234–249. - PubMed
-
- Cunnac S, Occhialini A, Barberis P, Boucher C, Genin S. Inventory and functional analysis of the large Hrp regulon in Ralstonia solanacearum: identification of novel effector proteins translocated to plant host cells through the type III secretion system. Mol Microbiol. 2004;53:115–128. - PubMed
-
- Tyler BM, Tripathy S, Zhang X, Dehal P, Jiang RH, et al. Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science. 2006;313:1261–1266. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

