On the edge of Bantu expansions: mtDNA, Y chromosome and lactase persistence genetic variation in southwestern Angola
- PMID: 19383166
- PMCID: PMC2682489
- DOI: 10.1186/1471-2148-9-80
On the edge of Bantu expansions: mtDNA, Y chromosome and lactase persistence genetic variation in southwestern Angola
Abstract
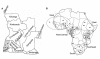
Background: Current information about the expansion of Bantu-speaking peoples is hampered by the scarcity of genetic data from well identified populations from southern Africa. Here, we fill an important gap in the analysis of the western edge of the Bantu migrations by studying for the first time the patterns of Y-chromosome, mtDNA and lactase persistence genetic variation in four representative groups living around the Namib Desert in southwestern Angola (Ovimbundu, Ganguela, Nyaneka-Nkumbi and Kuvale). We assessed the differentiation between these populations and their levels of admixture with Khoe-San groups, and examined their relationship with other sub-Saharan populations. We further combined our dataset with previously published data on Y-chromosome and mtDNA variation to explore a general isolation with migration model and infer the demographic parameters underlying current genetic diversity in Bantu populations.
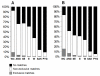
Results: Correspondence analysis, lineage sharing patterns and admixture estimates indicate that the gene pool from southwestern Angola is predominantly derived from West-Central Africa. The pastoralist Herero-speaking Kuvale people were additionally characterized by relatively high frequencies of Y-chromosome (12%) and mtDNA (22%) Khoe-San lineages, as well as by the presence of the -14010C lactase persistence mutation (6%), which likely originated in non-Bantu pastoralists from East Africa. Inferred demographic parameters show that both male and female populations underwent significant size growth after the split between the western and eastern branches of Bantu expansions occurring 4000 years ago. However, males had lower population sizes and migration rates than females throughout the Bantu dispersals.
Conclusion: Genetic variation in southwestern Angola essentially results from the encounter of an offshoot of West-Central Africa with autochthonous Khoisan-speaking peoples from the south. Interactions between the Bantus and the Khoe-San likely involved cattle herders from the two groups sharing common aspects of their social organization. The presence of the -14010C mutation in southwestern Angola provides a link between the East and Southwest African pastoral scenes that might have been established indirectly, through migrations of Khoe herders across southern Africa. Differences in patterns of mtDNA and Y-chromosome intrapopulation diversity and interpopulation differentiation may be explained by contrasting demographic histories underlying the current female and male genetic variation.
Figures




References
-
- Curtin P, Feierman S, Thompson L, Vansina J. African History: From Earliest Times to Independence. London, Longman; 1995.
-
- Newman JL. The peopling of Africa: A Geographical Interpretation. New Haven, Yale University Press; 1995.
-
- Ehret C. An African Classical Age: Eastern & Southern Africa in World History, 1000BC to AD400. Charlottesville, University Press of Virginia; 1998.
-
- Blench R. Archeology, Language, and the African Past. Lanham, Altamira Press; 2006.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

