New insights into the functional mechanisms and clinical applications of the kallikrein-related peptidase family
- PMID: 19383303
- PMCID: PMC5543873
- DOI: 10.1016/j.molonc.2007.09.003
New insights into the functional mechanisms and clinical applications of the kallikrein-related peptidase family
Abstract
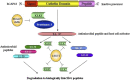
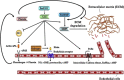
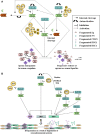
The Kallikrein-related peptidase (KLK) family consists of fifteen conserved serine proteases that form the largest contiguous cluster of proteases in the human genome. While primarily recognized for their clinical utilities as potential disease biomarkers, new compelling evidence suggests that this family plays a significant role in various physiological processes, including skin desquamation, semen liquefaction, neural plasticity, and body fluid homeostasis. KLK activation is believed to be mediated through highly organized proteolytic cascades, regulated through a series of feedback loops, inhibitors, auto-degradation and internal cleavages. Gene expression is mainly hormone-dependent, even though transcriptional epigenetic regulation has also been reported. These regulatory mechanisms are integrated with various signaling pathways to mediate multiple functions. Dysregulation of these pathways has been implicated in a large number of neoplastic and non-neoplastic pathological conditions. This review highlights our current knowledge of structural/phylogenetic features, functional role and regulatory/signaling mechanisms of this important family of enzymes.
Figures


References
-
- Amour, A. , Bird, M. , Chaudry, L. , Deadman, J. , Hayes, D. , Kay, C. , 2004. General considerations for proteolytic cascades. Biochem. Soc. Trans.. 32, 15–16. - PubMed
-
- Angelo, P.F. , Lima, A.R. , Alves, F.M. , Blaber, S.I. , Scarisbrick, I.A. , Blaber, M. , Juliano, L. , Juliano, M.A. , 2006. Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects. J. Biol. Chem.. 281, 3116–3126. - PubMed
-
- Bakin, R.E. , Gioeli, D. , Sikes, R.A. , Bissonette, E.A. , Weber, M.J. , 2003. Constitutive activation of the Ras/mitogen-activated protein kinase signaling pathway promotes androgen hypersensitivity in LNCaP prostate cancer cells. Cancer Res.. 63, 1981–1989. - PubMed
-
- Bando, Y. , Ito, S. , Nagai, Y. , Terayama, R. , Kishibe, M. , Jiang, Y.P. , Mitrovic, B. , Takahashi, T. , Yoshida, S. , 2006. Implications of protease M/neurosin in myelination during experimental demyelination and remyelination. Neurosci. Lett.. 405, 175–180. - PubMed
-
- Barrett, A.J. , Rawlings, N.D. , Woessner, J.F. , 2004. Handbook of Proteolytic Enzymes Academic Press Inc; San Diego:
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

