Aberrations of the MRE11-RAD50-NBS1 DNA damage sensor complex in human breast cancer: MRE11 as a candidate familial cancer-predisposing gene
- PMID: 19383352
- PMCID: PMC5527773
- DOI: 10.1016/j.molonc.2008.09.007
Aberrations of the MRE11-RAD50-NBS1 DNA damage sensor complex in human breast cancer: MRE11 as a candidate familial cancer-predisposing gene
Abstract
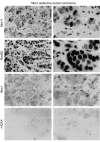
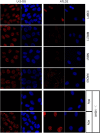
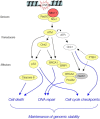
The MRE11, RAD50, and NBS1 genes encode proteins of the MRE11-RAD50-NBS1 (MRN) complex critical for proper maintenance of genomic integrity and tumour suppression; however, the extent and impact of their cancer-predisposing defects, and potential clinical value remain to be determined. Here, we report that among a large series of approximately 1000 breast carcinomas, around 3%, 7% and 10% tumours showed aberrantly reduced protein expression for RAD50, MRE11 and NBS1, respectively. Such defects were more frequent among the ER/PR/ERBB2 triple-negative and higher-grade tumours, among familial (especially BRCA1/BRCA2-associated) rather than sporadic cases, and the NBS1 defects correlated with shorter patients' survival. The BRCA1-associated and ER/PR/ERBB2 triple-negative tumours also showed high incidence of constitutively active DNA damage signalling (gammaH2AX) and p53 aberrations. Sequencing the RAD50, MRE11 and NBS1 genes of 8 patients from non-BRCA1/2 breast cancer families whose tumours showed concomitant reduction/loss of all three MRN-complex proteins revealed two germline mutations in MRE11: a missense mutation R202G and a truncating mutation R633STOP (R633X). Gene transfer and protein analysis of cell culture models with mutant MRE11 implicated various destabilization patterns among the MRN complex proteins including NBS1, the abundance of which was restored by re-expression of wild-type MRE11. We propose that germline mutations qualify MRE11 as a novel candidate breast cancer susceptibility gene in a subset of non-BRCA1/2 families. Our data have implications for the concept of the DNA damage response as an intrinsic anti-cancer barrier, various components of which become inactivated during cancer progression and also represent the bulk of breast cancer susceptibility genes discovered to date.
Figures






Similar articles
-
Expression of ATM, p53, and the MRE11-Rad50-NBS1 complex in myoepithelial cells from benign and malignant proliferations of the breast.J Clin Pathol. 2004 Nov;57(11):1179-84. doi: 10.1136/jcp.2004.017434. J Clin Pathol. 2004. PMID: 15509680 Free PMC article.
-
Breast cancer risk is associated with the genes encoding the DNA double-strand break repair Mre11/Rad50/Nbs1 complex.Cancer Epidemiol Biomarkers Prev. 2007 Oct;16(10):2024-32. doi: 10.1158/1055-9965.EPI-07-0116. Cancer Epidemiol Biomarkers Prev. 2007. PMID: 17932350
-
Rare key functional domain missense substitutions in MRE11A, RAD50, and NBN contribute to breast cancer susceptibility: results from a Breast Cancer Family Registry case-control mutation-screening study.Breast Cancer Res. 2014 Jun 3;16(3):R58. doi: 10.1186/bcr3669. Breast Cancer Res. 2014. PMID: 24894818 Free PMC article.
-
MRN (MRE11-RAD50-NBS1) Complex in Human Cancer and Prognostic Implications in Colorectal Cancer.Int J Mol Sci. 2019 Feb 14;20(4):816. doi: 10.3390/ijms20040816. Int J Mol Sci. 2019. PMID: 30769804 Free PMC article. Review.
-
The importance of making ends meet: mutations in genes and altered expression of proteins of the MRN complex and cancer.Mutat Res. 2008 Sep-Oct;659(3):262-73. doi: 10.1016/j.mrrev.2008.05.005. Epub 2008 Jun 23. Mutat Res. 2008. PMID: 18606567 Review.
Cited by
-
Genes associated with recurrence of hepatocellular carcinoma: integrated analysis by gene expression and methylation profiling.J Korean Med Sci. 2011 Nov;26(11):1428-38. doi: 10.3346/jkms.2011.26.11.1428. Epub 2011 Oct 27. J Korean Med Sci. 2011. PMID: 22065898 Free PMC article.
-
Suppression of Akt-mTOR pathway-a novel component of oncogene induced DNA damage response barrier in breast tumorigenesis.PLoS One. 2014 May 8;9(5):e97076. doi: 10.1371/journal.pone.0097076. eCollection 2014. PLoS One. 2014. PMID: 24811059 Free PMC article.
-
The Changing Landscape of Genetic Testing for Inherited Breast Cancer Predisposition.Curr Treat Options Oncol. 2017 May;18(5):27. doi: 10.1007/s11864-017-0468-y. Curr Treat Options Oncol. 2017. PMID: 28439798 Review.
-
DNA repair genes implicated in triple negative familial non-BRCA1/2 breast cancer predisposition.Am J Cancer Res. 2015 Jun 15;5(7):2113-26. eCollection 2015. Am J Cancer Res. 2015. PMID: 26328243 Free PMC article.
-
Heterozygous germline mutations in NBS1 among Korean patients with high-risk breast cancer negative for BRCA1/2 mutation.Fam Cancer. 2015 Sep;14(3):365-71. doi: 10.1007/s10689-015-9789-9. Fam Cancer. 2015. PMID: 25712764
References
-
- Aagaard, L. , Lukas, J. , Bartkova, J. , Kjerulff, A.A. , Strauss, M. , Bartek, J. , 1995. Aberrations of p16Ink4 and retinoblastoma tumour-suppressor genes occur in distinct sub-sets of human cancer cell lines. Int. J. Cancer 61, 115–120. - PubMed
-
- Ahmed, M. , Rahman, N. , 2006. ATM and breast cancer susceptibility. Oncogene 25, 5906–5911. - PubMed
-
- Angele, S. , Treilleux, I. , Bremond, A. , Taniere, P. , Hall, J. , 2003. Altered expression of DNA double-strand break detection and repair proteins in breast carcinomas. Histopathology 43, 347–353. - PubMed
-
- Bartek, J. , Bartkova, J. , Vojtesek, B. , Staskova, Z. , Lukas, J. , Rejthar, A. , Kovarik, J. , Midgley, C.M. , Gannon, J.V. , Lane, D.P. , 1991. Aberrant expression of the p53 oncoprotein is a common feature of a wide spectrum of human malignancies. Oncogene 6, 1699–1703. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

