The CALM and CALM/AF10 interactor CATS is a marker for proliferation
- PMID: 19383357
- PMCID: PMC5527775
- DOI: 10.1016/j.molonc.2008.08.001
The CALM and CALM/AF10 interactor CATS is a marker for proliferation
Abstract
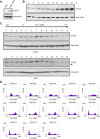
The CATS protein was recently identified as a novel CALM interacting protein. CATS increases the nuclear and specifically the nucleolar localization of the leukemogenic CALM/AF10 fusion protein. We cloned and characterized the murine Cats gene. Detailed analysis of murine Cats expression during mouse embryogenesis showed an association with rapidly proliferating tissues. Interestingly, the Cats transcript is highly expressed in murine hematopoietic cells transformed by CALM/AF10. The CATS protein is highly expressed in leukemia, lymphoma and tumor cell lines but not in non-proliferating T-cells or human peripheral blood lymphocytes. CATS protein levels are cell cycle dependent and it is induced by mitogens, suggesting a role of CATS in the control of cell proliferation and possibly CALM/AF10-mediated leukemogenesis.
Figures







Similar articles
-
The novel CALM interactor CATS influences the subcellular localization of the leukemogenic fusion protein CALM/AF10.Oncogene. 2006 Jul 6;25(29):4099-109. doi: 10.1038/sj.onc.1209438. Epub 2006 Feb 20. Oncogene. 2006. PMID: 16491119
-
A CALM-derived nuclear export signal is essential for CALM-AF10-mediated leukemogenesis.Blood. 2013 Jun 6;121(23):4758-68. doi: 10.1182/blood-2012-06-435792. Epub 2013 Mar 13. Blood. 2013. PMID: 23487024 Free PMC article.
-
The leukemogenic CALM/AF10 fusion protein alters the subcellular localization of the lymphoid regulator Ikaros.Oncogene. 2008 May 1;27(20):2886-96. doi: 10.1038/sj.onc.1210945. Epub 2007 Nov 26. Oncogene. 2008. PMID: 18037964
-
Clathrin assembly lymphoid myeloid leukemia-AF10-positive acute leukemias: a report of 2 cases with a review of the literature.Korean J Lab Med. 2010 Apr;30(2):117-21. doi: 10.3343/kjlm.2010.30.2.117. Korean J Lab Med. 2010. PMID: 20445327 Review.
-
The role of CALM-AF10 gene fusion in acute leukemia.Leukemia. 2008 Apr;22(4):678-85. doi: 10.1038/sj.leu.2405074. Epub 2007 Dec 20. Leukemia. 2008. PMID: 18094714 Free PMC article. Review.
Cited by
-
FAM64A contributes to ovarian cancer proliferation and metastasis by suppressing TWIST1 ubiquitination and degradation.Endocr Relat Cancer. 2025 Jun 16;32(6):e240048. doi: 10.1530/ERC-24-0048. Print 2025 Jun 1. Endocr Relat Cancer. 2025. PMID: 40424038 Free PMC article.
-
Prognostic implications of immune-related eight-gene signature in pediatric brain tumors.Braz J Med Biol Res. 2021 May 17;54(7):e10612. doi: 10.1590/1414-431X2020e10612. eCollection 2021. Braz J Med Biol Res. 2021. PMID: 34008756 Free PMC article. Clinical Trial.
-
Association of dietary folate and vitamin B-12 intake with genome-wide DNA methylation in blood: a large-scale epigenome-wide association analysis in 5841 individuals.Am J Clin Nutr. 2019 Aug 1;110(2):437-450. doi: 10.1093/ajcn/nqz031. Am J Clin Nutr. 2019. PMID: 31165884 Free PMC article.
-
FAM64A Promotes Osteosarcoma Cell Growth and Metastasis and Is Mediated by miR-493.J Oncol. 2020 Feb 24;2020:2518297. doi: 10.1155/2020/2518297. eCollection 2020. J Oncol. 2020. PMID: 32184824 Free PMC article.
-
FAM64A: A Novel Oncogenic Target of Lung Adenocarcinoma Regulated by Both Strands of miR-99a (miR-99a-5p and miR-99a-3p).Cells. 2020 Sep 11;9(9):2083. doi: 10.3390/cells9092083. Cells. 2020. PMID: 32932948 Free PMC article.
References
-
- Archangelo, L.F. , Glasner, J. , Krause, A. , Bohlander, S.K. , 2006. The novel CALM interactor CATS influences the subcellular localization of the leukemogenic fusion protein CALM/AF10. Oncogene 25, (29) 4099–4109. - PubMed
-
- Asnafi, V. , Radford-Weiss, I. , Dastugue, N. , Bayle, C. , Leboeuf, D. , Charrin, C. , Garand, R. , Lafage-Pochitaloff, M. , Delabesse, E. , Buzyn, A. , 2003. CALM-AF10 is a common fusion transcript in T-ALL and is specific to the TCRgammadelta lineage. Blood 102, (3) 1000–1006. - PubMed
-
- Asnafi, V. , Beldjord, K. , Libura, M. , Villarese, P. , Millien, C. , Ballerini, P. , Kuhlein, E. , Lafage-Pochitaloff, M. , Delabesse, E. , Bernard, O. , 2004. Age-related phenotypic and oncogenic differences in T-cell acute lymphoblastic leukemias may reflect thymic atrophy. Blood 104, (13) 4173–4180. - PubMed
-
- Asnafi, V. , Buzyn, A. , Thomas, X. , Huguet, F. , Vey, N. , Boiron, J.M. , Reman, O. , Cayuela, J.M. , Lheritier, V. , Vernant, J.P. , 2005. Impact of TCR status and genotype on outcome in adult T-cell acute lymphoblastic leukemia: a LALA-94 study. Blood 105, (8) 3072–3078. - PubMed
-
- Ayton, P.M. , Cleary, M.L. , 2001. Molecular mechanisms of leukemogenesis mediated by MLL fusion proteins. Oncogene 20, (40) 5695–5707. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

