Targeted proteomic strategy for clinical biomarker discovery
- PMID: 19383365
- PMCID: PMC2753590
- DOI: 10.1016/j.molonc.2008.12.001
Targeted proteomic strategy for clinical biomarker discovery
Abstract
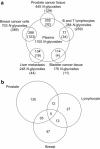
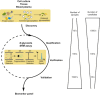
The high complexity and large dynamic range of blood plasma proteins currently prohibit the sensitive and high-throughput profiling of disease and control plasma proteome sample sets large enough to reliably detect disease indicating differences. To circumvent these technological limitations we describe here a new two-stage strategy for the mass spectrometry (MS) assisted discovery, verification and validation of disease biomarkers. In an initial discovery phase N-linked glycoproteins with distinguishable expression patterns in primary normal and diseased tissue are detected and identified. In the second step the proteins identified in the initial phase are subjected to targeted MS analysis in plasma samples, using the highly sensitive and specific selected reaction monitoring (SRM) technology. Since glycosylated proteins, such as those secreted or shed from the cell surface are likely to reside and persist in blood, the two-stage strategy is focused on the quantification of tissue derived glycoproteins in plasma. The focus on the N-glycoproteome not only reduces the complexity of the analytes, but also targets an information-rich subproteome which is relevant for remote sensing of diseases in the plasma. The N-glycoprotein based biomarker discovery and validation workflow reviewed here allows for the robust identification of protein candidate panels that can finally be selectively monitored in the blood plasma at high sensitivity in a reliable, non-invasive and quantitative fashion.
Figures

References
-
- Anderson, N.L. , Anderson, N.G. , 2002. The human plasma proteome: history, character, and diagnostic prospects. Mol. Cell. Proteomics. 1, 845–867. - PubMed
-
- Bantscheff, M. , Schirle, M. , Sweetman, G. , Rick, J. , Kuster, B. , 2007. Quantitative mass spectrometry in proteomics: a critical review. Anal. Bioanal. Chem.. 389, 1017–1031. - PubMed
-
- Baselga, J. , Norton, L. , Albanell, J. , Kim, Y.M. , Mendelsohn, J. , 1998. Recombinant humanized anti-HER2 antibody (Herceptin) enhances the antitumor activity of paclitaxel and doxorubicin against HER2/neu overexpressing human breast cancer xenografts. Cancer Res.. 58, 2825–2831. - PubMed
-
- Bayer, E.A. , Ben-Hur, H. , Wilchek, M. , 1988. Biocytin hydrazide–a selective label for sialic acids, galactose, and other sugars in glycoconjugates using avidin-biotin technology. Anal. Biochem.. 170, 271–281. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

