A C-terminal silencing domain in GW182 is essential for miRNA function
- PMID: 19383769
- PMCID: PMC2685512
- DOI: 10.1261/rna.1605509
A C-terminal silencing domain in GW182 is essential for miRNA function
Abstract
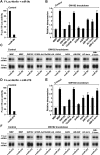
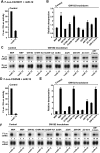
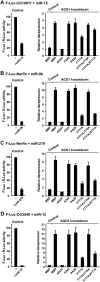
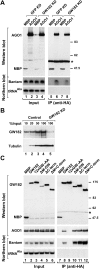
Proteins of the GW182 family are essential for miRNA-mediated gene silencing in animal cells; they interact with Argonaute proteins (AGOs) and are required for both the translational repression and mRNA degradation mediated by miRNAs. To gain insight into the role of the GW182-AGO1 interaction in silencing, we generated protein mutants that do not interact and tested them in complementation assays. We show that silencing of miRNA targets requires the N-terminal domain of GW182, which interacts with AGO1 through multiple glycine-tryptophan (GW)-repeats. Indeed, a GW182 mutant that does not interact with AGO1 cannot rescue silencing in cells depleted of endogenous GW182. Conversely, silencing is impaired by mutations in AGO1 that strongly reduce the interaction with GW182 but not with miRNAs. We further show that a GW182 mutant that does not localize to P-bodies but interacts with AGO1 rescues silencing in GW182-depleted cells, even though in these cells, AGO1 also fails to localize to P-bodies. Finally, we show that in addition to the N-terminal AGO1-binding domain, the middle and C-terminal regions of GW182 (referred to as the bipartite silencing domain) are essential for silencing. Together our results indicate that miRNA silencing in animal cells is mediated by AGO1 in complex with GW182, and that P-body localization is not required for silencing.
Figures


References
-
- Bagga S., Bracht J., Hunter S., Massirer K., Holtz J., Eachus R., Pasquinelli A.E. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. - PubMed
-
- Behm-Ansmant I., Rehwinkel J., Izaurralde E. MicroRNAs silence gene expression by repressing protein expression and/or by promoting mRNA decay. Cold Spring Harb. Symp. Quant. Biol. 2006b;71:523–530. - PubMed
-
- Bhattacharyya S.N., Habermacher R., Martine U., Closs E.I., Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
