Analysis of DNA copy number alterations in ovarian serous tumors identifies new molecular genetic changes in low-grade and high-grade carcinomas
- PMID: 19383911
- PMCID: PMC2782554
- DOI: 10.1158/0008-5472.CAN-08-3913
Analysis of DNA copy number alterations in ovarian serous tumors identifies new molecular genetic changes in low-grade and high-grade carcinomas
Erratum in
- Cancer Res. 2009 Jun 15;69(12):5267
Abstract
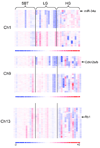
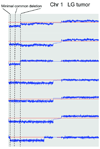
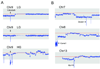
Ovarian serous carcinoma, the most common and lethal type of ovarian cancer, is thought to develop from two distinct molecular pathways. High-grade (HG) serous carcinomas contain frequent TP53 mutations, whereas low-grade (LG) carcinomas arise from serous borderline tumors (SBT) and harbor mutations in KRAS/BRAF/ERBB2 pathway. However, the molecular alterations involved in the progression from SBT to LG carcinoma remain unknown. In addition, the extent of deletion of tumor suppressors in ovarian serous carcinomas has not been well studied. To further address these two issues, we assessed DNA copy number changes among affinity-purified tumor cells from 37 ovarian serous neoplasms including SBT, LG, and HG tumors using high-density 250K single nucleotide polymorphism arrays. Chromosomal instability index as measured by changes in DNA copy number was significantly higher in HG than in LG serous carcinomas. Hemizygous ch1p36 deletion was common in LG serous carcinomas but was rarely seen in SBT. This region contains several candidate tumor suppressors including miR-34a. In contrast, in HG serous carcinomas, significant numbers of amplifications and deletions, including homozygous deletions, were identified. Among homozygous deletions, loci containing Rb1, CDKN2A/B, CSMD1, and DOCK4 were most common, being present in 10.6%, 6.4%, 6.4%, and 4.3%, respectively, in independent 47 affinity-purified HG serous carcinomas. Except for the CDKN2A/B region, these homozygous deletions were not present in either SBT or LG tumors. Our study provides a genome-wide homozygous deletion profile in HG serous carcinomas, which can serve as a molecular foundation to study tumor suppressors in ovarian cancer.
Figures



References
-
- Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. - PubMed
-
- Link CJ, Jr, Kohn E, Reed E. The relationship between borderline ovarian tumors and epithelial ovarian carcinoma: epidemiologic, pathologic, and molecular aspects. Gynecol Oncol. 1996;60:347–354. - PubMed
-
- Shih Ie M, Kurman RJ. Molecular pathogenesis of ovarian borderline tumors: new insights and old challenges. Clin Cancer Res. 2005;11:7273–7279. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

