Long-range epigenetic silencing at 2q14.2 affects most human colorectal cancers and may have application as a non-invasive biomarker of disease
- PMID: 19384295
- PMCID: PMC2696749
- DOI: 10.1038/sj.bjc.6605045
Long-range epigenetic silencing at 2q14.2 affects most human colorectal cancers and may have application as a non-invasive biomarker of disease
Abstract
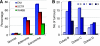
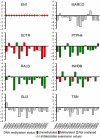
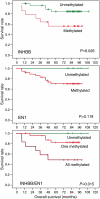
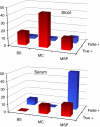
Large chromosomal regions can be suppressed in cancer cells as denoted by hypermethylation of neighbouring CpG islands and downregulation of most genes within the region. We have analysed the extent and prevalence of long-range epigenetic silencing at 2q14.2 (the first and best characterised example of coordinated epigenetic remodelling) and investigated its possible applicability as a non-invasive diagnostic marker of human colorectal cancer using different approaches and biological samples. Hypermethylation of at least one of the CpG islands analysed (EN1, SCTR, INHBB) occurred in most carcinomas (90%), with EN1 methylated in 73 and 40% of carcinomas and adenomas, respectively. Gene suppression was a common phenomenon in all the tumours analysed and affected both methylated and unmethylated genes. Detection of methylated EN1 using bisulfite treatment and melting curve (MC) analysis from stool DNA in patients and controls resulted in a predictive capacity of, 44% sensitivity in positive patients (27% of overall sensitivity) and 97% specificity. We conclude that epigenetic suppression along 2q14.2 is common to most colorectal cancers and the presence of a methylated EN1 CpG island in stool DNA might be used as biomarker of neoplastic disease.
Figures
References
-
- Ahlquist DA (2002) Stool-based DNA tests for colorectal cancer: clinical potential and early results. Rev Gastroenterol Disord 2(Suppl 1): S20–S26 - PubMed
-
- Baylin SB (2005) DNA methylation and gene silencing in cancer. Nat Clin Pract Oncol 2(Suppl 1): S4–S11 - PubMed
-
- Belshaw NJ, Elliott GO, Williams EA, Bradburn DM, Mills SJ, Mathers JC, Johnson IT (2004) Use of DNA from human stools to detect aberrant CpG island methylation of genes implicated in colorectal cancer. Cancer Epidemiol Biomarkers Prev 13: 1495–1501 - PubMed
-
- Brenner DE, Rennert G (2005) Fecal DNA biomarkers for the detection of colorectal neoplasia: attractive, but is it feasible? J Natl Cancer Inst 97: 1107–1109 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

