Rpb1 sumoylation in response to UV radiation or transcriptional impairment in yeast
- PMID: 19384408
- PMCID: PMC2668072
- DOI: 10.1371/journal.pone.0005267
Rpb1 sumoylation in response to UV radiation or transcriptional impairment in yeast
Abstract
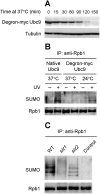
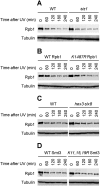
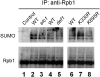
Covalent modifications of proteins by ubiquitin and the Small Ubiquitin-like MOdifier (SUMO) have been revealed to be involved in a plethora of cellular processes, including transcription, DNA repair and DNA damage responses. It has been well known that in response to DNA damage that blocks transcription elongation, Rpb1, the largest subunit of RNA polymerase II (Pol II), is ubiquitylated and subsequently degraded in mammalian and yeast cells. However, it is still an enigma regarding how Pol II responds to damaged DNA and conveys signal(s) for DNA damage-related cellular processes. We found that Rpb1 is also sumoylated in yeast cells upon UV radiation or impairment of transcription elongation, and this modification is independent of DNA damage checkpoint activation. Ubc9, an E2 SUMO conjugase, and Siz1, an E3 SUMO ligase, play important roles in Rpb1 sumoylation. K1487, which is located in the acidic linker region between the C-terminal domain and the globular domain of Rpb1, is the major sumoylation site. Rpb1 sumoylation is not affected by its ubiquitylation, and vice versa, indicating that the two processes do not crosstalk. Abolishment of Rpb1 sumoylation at K1487 does not affect transcription elongation or transcription coupled repair (TCR) of UV-induced DNA damage. However, deficiency in TCR enhances UV-induced Rpb1 sumoylation, presumably due to the persistence of transcription-blocking DNA lesions in the transcribed strand of a gene. Remarkably, abolishment of Rpb1 sumoylation at K1487 causes enhanced and prolonged UV-induced phosphorylation of Rad53, especially in TCR-deficient cells, suggesting that the sumoylation plays a role in restraining the DNA damage checkpoint response caused by transcription-blocking lesions. Our results demonstrate a novel covalent modification of Rpb1 in response to UV induced DNA damage or transcriptional impairment, and unravel an important link between the modification and the DNA damage checkpoint response.
Conflict of interest statement
Figures




References
-
- Ljungman M, Lane DP. Transcription - guarding the genome by sensing DNA damage. Nat Rev Cancer. 2004;4:727–737. - PubMed
-
- Harrison JC, Haber JE. Surviving the breakup: the DNA damage checkpoint. Annu Rev Genet. 2006;40:209–235. - PubMed
-
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, et al. DNA Repair and Mutagenesis. Washington D.C.: ASM Press; 2006.
-
- Verhage R, Zeeman AM, de Groot N, Gleig F, Bang DD, et al. The RAD7 and RAD16 genes, which are essential for pyrimidine dimer removal from the silent mating type loci, are also required for repair of the nontranscribed strand of an active gene in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:6135–6142. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

