The ordered extension of pseudopodia by amoeboid cells in the absence of external cues
- PMID: 19384419
- PMCID: PMC2668753
- DOI: 10.1371/journal.pone.0005253
The ordered extension of pseudopodia by amoeboid cells in the absence of external cues
Abstract
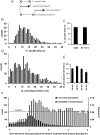
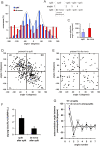
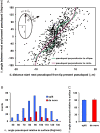
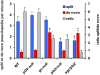
Eukaryotic cells extend pseudopodia for movement. In the absence of external cues, cells move in random directions, but with a strong element of persistence that keeps them moving in the same direction Persistence allows cells to disperse over larger areas and is instrumental to enter new environments where spatial cues can lead the cell. Here we explore cell movement by analyzing the direction, size and timing of approximately 2000 pseudopodia that are extended by Dictyostelium cells. The results show that pseudpopod are extended perpendicular to the surface curvature at the place where they emerge. The location of new pseudopods is not random but highly ordered. Two types of pseudopodia may be formed: frequent splitting of an existing pseudopod, or the occasional extension of a de novo pseudopod at regions devoid of recent pseudopod activity. Split-pseudopodia are extended at approximately 60 degrees relative to the previous pseudopod, mostly as alternating Right/Left/Right steps leading to relatively straight zigzag runs. De novo pseudopodia are extended in nearly random directions thereby interrupting the zigzag runs. Persistence of cell movement is based on the ratio of split versus de novo pseudopodia. We identify PLA2 and cGMP signaling pathways that modulate this ratio of splitting and de novo pseudopodia, and thereby regulate the dispersal of cells. The observed ordered extension of pseudopodia in the absence of external cues provides a fundamental insight into the coordinated movement of cells, and might form the basis for movement that is directed by internal or external cues.
Conflict of interest statement
Figures


References
-
- Pollard TD. The cytoskeleton, cellular motility and the reductionist agenda. Nature. 2003;422:741–745. - PubMed
-
- Van Haastert PJM, Devreotes PN. Chemotaxis: signalling the way forward. Nat Rev Mol Cell Biol. 2004;5:626–634. - PubMed
-
- Patlak CS. Random walk with persistence and external bias. Bull Math Biophys. 1953;15:311–338.
-
- Potel MJ, Mackay SA. Preaggregative cell motion in Dictyostelium. J Cell Sci. 1979;36:281–309. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

