Impact of pretreated Switchgrass and biomass carbohydrates on Clostridium thermocellum ATCC 27405 cellulosome composition: a quantitative proteomic analysis
- PMID: 19384422
- PMCID: PMC2668762
- DOI: 10.1371/journal.pone.0005271
Impact of pretreated Switchgrass and biomass carbohydrates on Clostridium thermocellum ATCC 27405 cellulosome composition: a quantitative proteomic analysis
Abstract
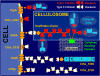
Background: Economic feasibility and sustainability of lignocellulosic ethanol production requires the development of robust microorganisms that can efficiently degrade and convert plant biomass to ethanol. The anaerobic thermophilic bacterium Clostridium thermocellum is a candidate microorganism as it is capable of hydrolyzing cellulose and fermenting the hydrolysis products to ethanol and other metabolites. C. thermocellum achieves efficient cellulose hydrolysis using multiprotein extracellular enzymatic complexes, termed cellulosomes.
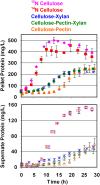
Methodology/principal findings: In this study, we used quantitative proteomics (multidimensional LC-MS/MS and (15)N-metabolic labeling) to measure relative changes in levels of cellulosomal subunit proteins (per CipA scaffoldin basis) when C. thermocellum ATCC 27405 was grown on a variety of carbon sources [dilute-acid pretreated switchgrass, cellobiose, amorphous cellulose, crystalline cellulose (Avicel) and combinations of crystalline cellulose with pectin or xylan or both]. Cellulosome samples isolated from cultures grown on these carbon sources were compared to (15)N labeled cellulosome samples isolated from crystalline cellulose-grown cultures. In total from all samples, proteomic analysis identified 59 dockerin- and 8 cohesin-module containing components, including 16 previously undetected cellulosomal subunits. Many cellulosomal components showed differential protein abundance in the presence of non-cellulose substrates in the growth medium. Cellulosome samples from amorphous cellulose, cellobiose and pretreated switchgrass-grown cultures displayed the most distinct differences in composition as compared to cellulosome samples from crystalline cellulose-grown cultures. While Glycoside Hydrolase Family 9 enzymes showed increased levels in the presence of crystalline cellulose, and pretreated switchgrass, in particular, GH5 enzymes showed increased levels in response to the presence of cellulose in general, amorphous or crystalline.
Conclusions/significance: Overall, the quantitative results suggest a coordinated substrate-specific regulation of cellulosomal subunit composition in C. thermocellum to better suit the organism's needs for growth under different conditions. To date, this study provides the most comprehensive comparison of cellulosomal compositional changes in C. thermocellum in response to different carbon sources. Such studies are vital to engineering a strain that is best suited to grow on specific substrates of interest and provide the building blocks for constructing designer cellulosomes with tailored enzyme composition for industrial ethanol production.
Conflict of interest statement
Figures


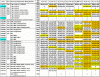

 indicates that the protein was identified in both 14N and 15N forms in both BR; A
indicates that the protein was identified in both 14N and 15N forms in both BR; A Similar articles
-
Clostridium clariflavum: Key Cellulosome Players Are Revealed by Proteomic Analysis.mBio. 2015 May 19;6(3):e00411-15. doi: 10.1128/mBio.00411-15. mBio. 2015. PMID: 25991683 Free PMC article.
-
How does cellulosome composition influence deconstruction of lignocellulosic substrates in Clostridium (Ruminiclostridium) thermocellum DSM 1313?Biotechnol Biofuels. 2017 Sep 18;10:222. doi: 10.1186/s13068-017-0909-7. eCollection 2017. Biotechnol Biofuels. 2017. PMID: 28932263 Free PMC article.
-
Stoichiometric Assembly of the Cellulosome Generates Maximum Synergy for the Degradation of Crystalline Cellulose, as Revealed by In Vitro Reconstitution of the Clostridium thermocellum Cellulosome.Appl Environ Microbiol. 2015 Jul;81(14):4756-66. doi: 10.1128/AEM.00772-15. Epub 2015 May 8. Appl Environ Microbiol. 2015. PMID: 25956772 Free PMC article.
-
[Mics of the Clostridium thermocellum in lignocellulose degradation--a review].Wei Sheng Wu Xue Bao. 2014 Feb 4;54(2):121-8. Wei Sheng Wu Xue Bao. 2014. PMID: 24818461 Review. Chinese.
-
Cellulosomes: Highly Efficient Cellulolytic Complexes.Subcell Biochem. 2021;96:323-354. doi: 10.1007/978-3-030-58971-4_9. Subcell Biochem. 2021. PMID: 33252735 Review.
Cited by
-
Proteomic analysis of Clostridium thermocellum core metabolism: relative protein expression profiles and growth phase-dependent changes in protein expression.BMC Microbiol. 2012 Sep 21;12:214. doi: 10.1186/1471-2180-12-214. BMC Microbiol. 2012. PMID: 22994686 Free PMC article.
-
Composition and yield of non-cellulosic and cellulosic sugars in soluble and particulate fractions during consolidated bioprocessing of poplar biomass by Clostridium thermocellum.Biotechnol Biofuels Bioprod. 2022 Feb 28;15(1):23. doi: 10.1186/s13068-022-02119-9. Biotechnol Biofuels Bioprod. 2022. PMID: 35227303 Free PMC article.
-
Clostridium thermocellum transcriptomic profiles after exposure to furfural or heat stress.Biotechnol Biofuels. 2013 Sep 12;6(1):131. doi: 10.1186/1754-6834-6-131. Biotechnol Biofuels. 2013. PMID: 24028713 Free PMC article.
-
The emergence of Clostridium thermocellum as a high utility candidate for consolidated bioprocessing applications.Front Chem. 2014 Aug 26;2:66. doi: 10.3389/fchem.2014.00066. eCollection 2014. Front Chem. 2014. PMID: 25207268 Free PMC article. Review.
-
Enzymatic diversity of the Clostridium thermocellum cellulosome is crucial for the degradation of crystalline cellulose and plant biomass.Sci Rep. 2016 Oct 19;6:35709. doi: 10.1038/srep35709. Sci Rep. 2016. PMID: 27759119 Free PMC article.
References
-
- Bayer EA, Shoham Y, Lamed R. Cellulose-decomposing bacteria and their enzyme systems. In: Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E, editors. The Prokaryotes: Ecophysiology and Biochemistry. New York: Springer; 2006. pp. 578–617.
-
- Bayer EA, Belaich JP, Shoham Y, Lamed R. The cellulosomes: multienzyme machines for degradation of plant cell wall polysaccharides. Annu Rev Microbiol. 2004;58:521–554. - PubMed
-
- Bayer EA, Chanzy H, Lamed R, Shoham Y. Cellulose, cellulases and cellulosomes. Curr Opin Struct Biol. 1998;8:548–557. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

