Practical protocols for production of very high yields of recombinant proteins using Escherichia coli
- PMID: 19384993
- PMCID: PMC2771296
- DOI: 10.1002/pro.102
Practical protocols for production of very high yields of recombinant proteins using Escherichia coli
Abstract
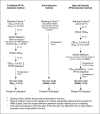
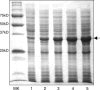
The gram-negative bacterium Escherichia coli offers a mean for rapid, high yield, and economical production of recombinant proteins. However, high-level production of functional eukaryotic proteins in E. coli may not be a routine matter, sometimes it is quite challenging. Techniques to optimize heterologous protein overproduction in E. coli have been explored for host strain selection, plasmid copy numbers, promoter selection, mRNA stability, and codon usage, significantly enhancing the yields of the foreign eukaryotic proteins. We have been working on optimizations of bacterial expression conditions and media with a focus on achieving very high cell density for high-level production of eukaryotic proteins. Two high-cell-density bacterial expression methods have been explored, including an autoinduction introduced by Studier (Protein Expr Purif 2005;41:207-234) recently and a high-cell-density IPTG-induction method described in this study, to achieve a cell-density OD(600) of 10-20 in the normal laboratory setting using a regular incubator shaker. Several practical protocols have been implemented with these high-cell-density expression methods to ensure a very high yield of recombinant protein production. With our methods and protocols, we routinely obtain 14-25 mg of NMR triple-labeled proteins and 17-34 mg of unlabeled proteins from a 50-mL cell culture for all seven proteins we tested. Such a high protein yield used the same DNA constructs, bacterial strains, and a regular incubator shaker and no fermentor is necessary. More importantly, these methods allow us to consistently obtain such a high yield of recombinant proteins using E. coli expression.
Figures

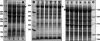



References
-
- Swartz JR. Advances in Escherichia coli production of therapeutic proteins. Curr Opin Biotechnol. 2001;12:195–201. - PubMed
-
- Hewitt L, McDonnell JM. Screening and optimizing protein production in E. coli methods. Mol Biol. 2004;278:1–16. - PubMed
-
- Jana S, Deb JK. Strategies for efficient production of heterologous proteins in Escherichia coli. Appl Microbiol Biotechnol. 2005;67:289–298. - PubMed
-
- Peti W, Page R. 2006
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

